Influence of solubilizing environments on membrane protein structures
- PMID: 20724162
- PMCID: PMC3161620
- DOI: 10.1016/j.tibs.2010.07.005
Influence of solubilizing environments on membrane protein structures
Abstract
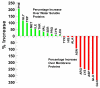
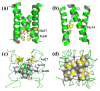
Membrane protein structures are stabilized by weak interactions and are influenced by additional interactions with the solubilizing environment. Structures of influenza virus A M2 protein, a proven drug target, have been determined in three different environments, thus providing a unique opportunity to assess environmental influences. Structures determined in detergents and detergent micelles can have notable differences from those determined in lipid bilayers. These differences make it imperative to validate membrane protein structures.
Copyright © 2010 Elsevier Ltd. All rights reserved.
Figures
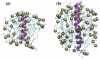

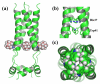

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

