Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1
- PMID: 20724438
- PMCID: PMC3017600
- DOI: 10.1093/nar/gkq743
Structure and function of the Rad9-binding region of the DNA-damage checkpoint adaptor TopBP1
Erratum in
- Nucleic Acids Res. 2013 Apr;41(8):4741
Abstract
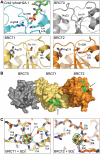
TopBP1 is a scaffold protein that coordinates activation of the DNA-damage-checkpoint response by coupling binding of the 9-1-1 checkpoint clamp at sites of ssDNA, to activation of the ATR-ATRIP checkpoint kinase complex. We have now determined the crystal structure of the N-terminal region of human TopBP1, revealing an unexpected triple-BRCT domain structure. The arrangement of the BRCT domains differs significantly from previously described tandem BRCT domain structures, and presents two distinct sites for binding phosphopeptides in the second and third BRCT domains. We show that the site in the second but not third BRCT domain in the N-terminus of TopBP1, provides specific interaction with a phosphorylated motif at pSer387 in Rad9, which can be generated by CK2.
Figures




Similar articles
-
BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.Elife. 2018 Oct 8;7:e39979. doi: 10.7554/eLife.39979. Elife. 2018. PMID: 30295604 Free PMC article.
-
Phosphorylation-dependent assembly and coordination of the DNA damage checkpoint apparatus by Rad4(TopBP1).Mol Cell. 2013 Sep 26;51(6):723-736. doi: 10.1016/j.molcel.2013.08.030. Mol Cell. 2013. PMID: 24074952 Free PMC article.
-
Interaction between Rad9-Hus1-Rad1 and TopBP1 activates ATR-ATRIP and promotes TopBP1 recruitment to sites of UV-damage.DNA Repair (Amst). 2014 Sep;21:1-11. doi: 10.1016/j.dnarep.2014.05.001. Epub 2014 May 27. DNA Repair (Amst). 2014. PMID: 25091155
-
TopBP1: A BRCT-scaffold protein functioning in multiple cellular pathways.DNA Repair (Amst). 2014 Oct;22:165-74. doi: 10.1016/j.dnarep.2014.06.004. Epub 2014 Jul 30. DNA Repair (Amst). 2014. PMID: 25087188 Review.
-
TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation.Cell Cycle. 2009 Sep 15;8(18):2877-84. doi: 10.4161/cc.8.18.9485. Epub 2009 Sep 9. Cell Cycle. 2009. PMID: 19652550 Review.
Cited by
-
Functional Evolution of BRCT Domains from Binding DNA to Protein.Evol Bioinform Online. 2011;7:87-97. doi: 10.4137/EBO.S7084. Epub 2011 Jun 12. Evol Bioinform Online. 2011. PMID: 21814458 Free PMC article.
-
ADAR1 links R-loop homeostasis to ATR activation in replication stress response.Nucleic Acids Res. 2023 Nov 27;51(21):11668-11687. doi: 10.1093/nar/gkad839. Nucleic Acids Res. 2023. PMID: 37831098 Free PMC article.
-
Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit.PLoS One. 2013 Dec 20;8(12):e85859. doi: 10.1371/journal.pone.0085859. eCollection 2013. PLoS One. 2013. PMID: 24376897 Free PMC article.
-
BRCT domains of the DNA damage checkpoint proteins TOPBP1/Rad4 display distinct specificities for phosphopeptide ligands.Elife. 2018 Oct 8;7:e39979. doi: 10.7554/eLife.39979. Elife. 2018. PMID: 30295604 Free PMC article.
-
Phosphorylation-dependent assembly of DNA damage response systems and the central roles of TOPBP1.DNA Repair (Amst). 2021 Dec;108:103232. doi: 10.1016/j.dnarep.2021.103232. Epub 2021 Sep 29. DNA Repair (Amst). 2021. PMID: 34678589 Free PMC article. Review.
References
-
- Smits VA, Warmerdam DO, Martin Y, Freire R. Mechanisms of ATR-mediated checkpoint signalling. Front. Biosci. 2010;15:840–853. - PubMed
-
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. - PubMed
-
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:227–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

