Upstream open reading frames: molecular switches in (patho)physiology
- PMID: 20726009
- PMCID: PMC3045505
- DOI: 10.1002/bies.201000037
Upstream open reading frames: molecular switches in (patho)physiology
Abstract
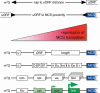
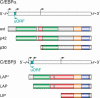
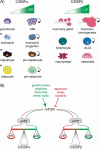
Conserved upstream open reading frames (uORFs) are found within many eukaryotic transcripts and are known to regulate protein translation. Evidence from genetic and bioinformatic studies implicates disturbed uORF-mediated translational control in the etiology of human diseases. A genetic mouse model has recently provided proof-of-principle support for the physiological relevance of uORF-mediated translational control in mammals. The targeted disruption of the uORF initiation codon within the transcription factor CCAAT/enhancer binding protein β (C/EBPβ) gene resulted in deregulated C/EBPβ protein isoform expression, associated with defective liver regeneration and impaired osteoclast differentiation. The high prevalence of uORFs in the human transcriptome suggests that intensified search for mutations within 5' RNA leader regions may reveal a multitude of alterations affecting uORFs, causing pathogenic deregulation of protein expression.
Copyright © 2010 WILEY Periodicals, Inc.
Figures

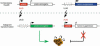
References
-
- Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–23. - PubMed
-
- Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin Cell Dev Biol. 2005;16:39–47. - PubMed
-
- Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem Sci. 2003;28:182–8. - PubMed
-
- Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell. 2009;101:251–62. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

