Isolation and characterization of the core single-stranded DNA-binding domain of purine-rich element binding protein B (Purβ)
- PMID: 20728429
- PMCID: PMC2957832
- DOI: 10.1016/j.bbrc.2010.08.059
Isolation and characterization of the core single-stranded DNA-binding domain of purine-rich element binding protein B (Purβ)
Abstract
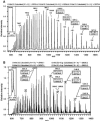
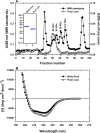
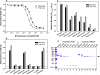
Purβ is a single-stranded nucleic acid-binding protein implicated in the injury-induced repression of genes encoding certain muscle-restricted isoforms of actin and myosin expressed in the heart, skeletal muscle, and vasculature. To better understand how the modular arrangement of the primary sequence of Purβ affects the higher order structure and function of the protein, purified recombinant Purβ was subjected to partial proteolysis in an attempt to identify a well-folded truncation protein that retained purine-rich single-stranded DNA-binding activity. Limited tryptic digestion of Purβ liberated a core ∼30kDa fragment corresponding to residues 29-305 as determined by epitope mapping and mass spectrometry. Size exclusion chromatography indicated that the isolated core fragment retains the ability to self-associate while circular dichroism analysis confirmed that the Purβ core domain is stably folded in the absence of glycine-rich N- and C-terminal sequences. Comparative DNA-binding assays revealed that the isolated core domain interacts with purine-rich cis-elements from the smooth muscle α-actin gene with similar specificity but increased affinity compared to full-length Purβ. These findings suggest that the highly conserved modular repeats of Purβ fold to form a core functional domain, which mediates the specific and high affinity binding of the protein to single-stranded DNA.
Copyright © 2010 Elsevier Inc. All rights reserved.
Figures

References
-
- Johnson EM. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res. 2003;23:2093–2100. - PubMed
-
- Kelm RJ, Jr., Cogan JG, Elder PK, Strauch AR, Getz MJ. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle α-actin promoter. J. Biol. Chem. 1999;274:14238–14245. - PubMed
-
- Carlini LE, Getz MJ, Strauch AR, Kelm RJ., Jr. Cryptic MCAT enhancer regulation in fibroblasts and smooth muscle cells. Suppression of TEF-1 mediated activation by the single-stranded DNA-binding proteins, Purα, Purβ, and MSY1. J. Biol. Chem. 2002;277:8682–8692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

