Chromatin regulation and genome maintenance by mammalian SIRT6
- PMID: 20729089
- PMCID: PMC2991557
- DOI: 10.1016/j.tibs.2010.07.009
Chromatin regulation and genome maintenance by mammalian SIRT6
Abstract
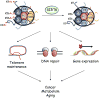
Saccharomyces cerevisiae Sir2 is an NAD(+)-dependent histone deacetylase that links chromatin silencing to genomic stability, cellular metabolism and lifespan regulation. In mice, deficiency for the Sir2 family member SIRT6 leads to genomic instability, metabolic defects and degenerative pathologies associated with aging. Until recently, SIRT6 was an orphan enzyme whose catalytic activity and substrates were unclear. However, new mechanistic insights have come from the discovery that SIRT6 is a highly substrate-specific histone deacetylase that promotes proper chromatin function in several physiologic contexts, including telomere and genome stabilization, gene expression and DNA repair. By maintaining both the integrity and the expression of the mammalian genome, SIRT6 thus serves several roles that parallel Sir2 function. In this article, we review recent advances in understanding the mechanisms of SIRT6 action and their implications for human biology and disease.
Published by Elsevier Ltd.
Figures


References
-
- Gottschling DE, et al. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. - PubMed
-
- Aparicio OM, et al. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. - PubMed
-
- Bryk M, et al. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. - PubMed
-
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

