Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis
- PMID: 20729191
- PMCID: PMC2963421
- DOI: 10.1074/jbc.M110.151316
Further insights into the roles of GTP and the C terminus of the hepatitis C virus polymerase in the initiation of RNA synthesis
Abstract
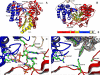
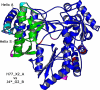
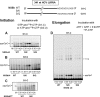
The hepatitis C virus (HCV) NS5b protein is an RNA-dependent RNA polymerase essential for replication of the viral RNA genome. In vitro and presumably in vivo, NS5b initiates RNA synthesis by a de novo mechanism. Different structural elements of NS5b have been reported to participate in RNA synthesis, especially a so-called "β-flap" and a C-terminal segment (designated "linker") that connects the catalytic core of NS5b to a transmembrane anchor. High concentrations of GTP have also been shown to stimulate de novo RNA synthesis by HCV NS5b. Here we describe a combined structural and functional analysis of genotype 1 HCV-NS5b of strains H77 (subtype 1a), for which no structure has been previously reported, and J4 (subtype 1b). Our results highlight the linker as directly involved in lifting the first boundary to processive RNA synthesis, the formation of the first dinucleotide primer. The transition from this first dinucleotide primer state to processive RNA synthesis requires removal of the linker and of the β-flap with which it is shown to strongly interact in crystal structures of HCV NS5b. We find that GTP specifically stimulates this transition irrespective of its incorporation in neosynthesized RNA.
Figures





Similar articles
-
Two crucial early steps in RNA synthesis by the hepatitis C virus polymerase involve a dual role of residue 405.J Virol. 2012 Jul;86(13):7107-17. doi: 10.1128/JVI.00459-12. Epub 2012 Apr 24. J Virol. 2012. PMID: 22532694 Free PMC article.
-
Functional characterization of fingers subdomain-specific monoclonal antibodies inhibiting the hepatitis C virus RNA-dependent RNA polymerase.J Biol Chem. 2008 Aug 29;283(35):24089-102. doi: 10.1074/jbc.M803422200. Epub 2008 Jun 23. J Biol Chem. 2008. PMID: 18574240 Free PMC article.
-
Selection of 3'-template bases and initiating nucleotides by hepatitis C virus NS5B RNA-dependent RNA polymerase.J Virol. 2002 Jul;76(14):7030-9. doi: 10.1128/jvi.76.14.7030-7039.2002. J Virol. 2002. PMID: 12072503 Free PMC article.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
Structures of hepatitis C virus nonstructural proteins required for replicase assembly and function.Curr Opin Virol. 2013 Apr;3(2):129-36. doi: 10.1016/j.coviro.2013.03.013. Epub 2013 Apr 16. Curr Opin Virol. 2013. PMID: 23601958 Free PMC article. Review.
Cited by
-
Viral Capsid and Polymerase in Reoviridae.Subcell Biochem. 2022;99:525-552. doi: 10.1007/978-3-031-00793-4_17. Subcell Biochem. 2022. PMID: 36151388
-
Mutations Identified in the Hepatitis C Virus (HCV) Polymerase of Patients with Chronic HCV Treated with Ribavirin Cause Resistance and Affect Viral Replication Fidelity.Antimicrob Agents Chemother. 2020 Nov 17;64(12):e01417-20. doi: 10.1128/AAC.01417-20. Print 2020 Nov 17. Antimicrob Agents Chemother. 2020. PMID: 32928732 Free PMC article. Clinical Trial.
-
Annotating Protein Functional Residues by Coupling High-Throughput Fitness Profile and Homologous-Structure Analysis.mBio. 2016 Nov 1;7(6):e01801-16. doi: 10.1128/mBio.01801-16. mBio. 2016. PMID: 27803181 Free PMC article.
-
Inhibitors of the Hepatitis C Virus RNA-Dependent RNA Polymerase NS5B.Viruses. 2010 Oct;2(10):2169-2195. doi: 10.3390/v2102169. Epub 2010 Sep 28. Viruses. 2010. PMID: 21994615 Free PMC article.
-
Bluetongue virus VP1 polymerase activity in vitro: template dependency, dinucleotide priming and cap dependency.PLoS One. 2011;6(11):e27702. doi: 10.1371/journal.pone.0027702. Epub 2011 Nov 15. PLoS One. 2011. PMID: 22110731 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

