An extreme thermophile, Thermus thermophilus, is a polyploid bacterium
- PMID: 20729360
- PMCID: PMC2950507
- DOI: 10.1128/JB.00662-10
An extreme thermophile, Thermus thermophilus, is a polyploid bacterium
Abstract
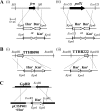
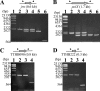
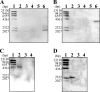
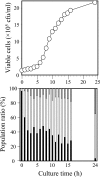
An extremely thermophilic bacterium, Thermus thermophilus HB8, is one of the model organisms for systems biology. Its genome consists of a chromosome (1.85 Mb), a megaplasmid (0.26 Mb) designated pTT27, and a plasmid (9.3 kb) designated pTT8, and the complete sequence is available. We show here that T. thermophilus is a polyploid organism, harboring multiple genomic copies in a cell. In the case of the HB8 strain, the copy number of the chromosome was estimated to be four or five, and the copy number of the pTT27 megaplasmid seemed to be equal to that of the chromosome. It has never been discussed whether T. thermophilus is haploid or polyploid. However, the finding that it is polyploid is not surprising, as Deinococcus radiodurans, an extremely radioresistant bacterium closely related to Thermus, is well known to be a polyploid organism. As is the case for D. radiodurans in the radiation environment, the polyploidy of T. thermophilus might allow for genomic DNA protection, maintenance, and repair at elevated growth temperatures. Polyploidy often complicates the recognition of an essential gene in T. thermophilus as a model organism for systems biology.
Figures
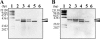


References
-
- Angert, E. R., and K. D. Clements. 2004. Initiation of intracellular offspring in Epulopiscium. Mol. Microbiol. 51:827-835. - PubMed
-
- Bremer, H., and P. P. Dennis. 1996. Modulation of chemical composition and other parameters of the cell growth rate, p. 1553-1566. In F. C. Neidhardt (ed.), Escherichia coli and Salmonella. ASM Press, Washington, DC.
-
- Brouns, S. J., H. Wu, J. Akerboom, A. P. Turnbull, W. M. de Vos, and J. van der Oost. 2005. Engineering a selectable marker for hyperthermophiles. J. Biol. Chem. 280:11422-11431. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

