Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria
- PMID: 20729366
- PMCID: PMC2950506
- DOI: 10.1128/JB.00592-10
Characterization of the RNA degradosome of Pseudoalteromonas haloplanktis: conservation of the RNase E-RhlB interaction in the gammaproteobacteria
Abstract
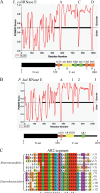
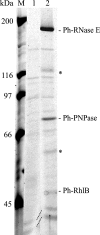
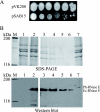
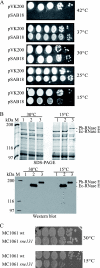
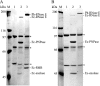
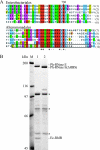
The degradosome is a multienzyme complex involved in mRNA degradation in Escherichia coli. The essential endoribonuclease RNase E contains a large noncatalytic region necessary for protein-protein interactions with other components of the RNA degradosome. Interacting proteins include the DEAD-box RNA helicase RhlB, the glycolytic enzyme enolase, and the exoribonuclease PNPase. Pseudoalteromonas haloplanktis, a psychrotolerant gammaproteobacterium distantly related to E. coli, encodes homologs of each component of the RNA degradosome. In P. haloplanktis, RNase E associates with RhlB and PNPase but not enolase. Plasmids expressing P. haloplanktis RNase E (Ph-RNase E) can complement E. coli strains lacking E. coli RNase E (Ec-RNase E). Ph-RNase E, however, does not confer a growth advantage to E. coli at low temperature. Ph-RNase E has a heterologous protein-protein interaction with Ec-RhlB but not with Ec-enolase or Ec-PNPase. The Ph-RNase E binding sites for RhlB and PNPase were mapped by deletion analysis. The PNPase binding site is located at the C-terminal end of Ph-RNase E at the same position as that in Ec-RNase E, but the sequence of the site is not conserved. The sequence of the RhlB binding site in Ph-RNase E is related to the sequence in Ec-RNase E. Together with the heterologous interaction between Ph-RNase E and Ec-RhlB, our results suggest that the underlying structural motif for the RNase E-RhlB interaction is conserved. Since the activity of Ec-RhlB requires its physical interaction with Ec-RNase E, conservation of the underlying structural motif over a large evolutionary distance could be due to constraints involved in the control of RhlB activity.
Figures

References
-
- Birolo, L., M. L. Tutino, B. Fontanella, C. Gerday, K. Mainolfi, S. Pascarella, G. Sannia, F. Vinci, and G. Marino. 2000. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC 125. Cloning, expression, properties, and molecular modelling. Eur. J. Biochem. 267:2790-2802. - PubMed
-
- Callaghan, A. J., J. P. Aurikko, L. L. Ilag, J. Gunter Grossmann, V. Chandran, K. Kuhnel, L. Poljak, A. J. Carpousis, C. V. Robinson, M. F. Symmons, and B. F. Luisi. 2004. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J. Mol. Biol. 340:965-979. - PubMed
-
- Callaghan, A. J., M. J. Marcaida, J. A. Stead, K. J. McDowall, W. G. Scott, and B. F. Luisi. 2005. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature 437:1187-1191. - PubMed
-
- Carpousis, A. J. 2002. The Escherichia coli RNA degradosome: structure, function and relationship to other ribonucleolytic multienzyme complexes. Biochem. Soc. Trans. 30:150-155. - PubMed
-
- Carpousis, A. J., V. Khemici, S. Ait-Bara, and L. Poljak. 2008. Co-immunopurification of multiprotein complexes containing RNA-degrading enzymes. Methods Enzymol. 447:65-82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

