The effect of electrostatics on the marginal cooperativity of an ultrafast folding protein
- PMID: 20729560
- PMCID: PMC2966070
- DOI: 10.1074/jbc.M110.154021
The effect of electrostatics on the marginal cooperativity of an ultrafast folding protein
Abstract
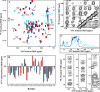
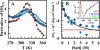
Proteins fold up by coordinating the different segments of their polypeptide chain through a network of weak cooperative interactions. Such cooperativity results in unfolding curves that are typically sigmoidal. However, we still do not know what factors modulate folding cooperativity or the minimal amount that ensures folding into specific three-dimensional structures. Here, we address these issues on BBL, a small helical protein that folds in microseconds via a marginally cooperative downhill process (Li, P., Oliva, F. Y., Naganathan, A. N., and Muñoz, V. (2009) Proc. Natl. Acad. Sci. USA. 106, 103-108). Particularly, we explore the effects of salt-induced screening of the electrostatic interactions in BBL at neutral pH and in acid-denatured BBL. Our results show that electrostatic screening stabilizes the native state of the neutral and protonated forms, inducing complete refolding of acid-denatured BBL. Furthermore, without net electrostatic interactions, the unfolding process becomes much less cooperative, as judged by the broadness of the equilibrium unfolding curve and the relaxation rate. Our experiments show that the marginally cooperative unfolding of BBL can still be made twice as broad while the protein retains its ability to fold into the native three-dimensional structure in microseconds. This result demonstrates experimentally that efficient folding does not require cooperativity, confirming predictions from theory and computer simulations and challenging the conventional biochemical paradigm. Furthermore, we conclude that electrostatic interactions are an important factor in determining folding cooperativity. Thus, electrostatic modulation by pH-salt and/or mutagenesis of charged residues emerges as an attractive tool for tuning folding cooperativity.
Figures

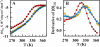

Similar articles
-
Downhill versus barrier-limited folding of BBL 3. Heterogeneity of the native state of the BBL peripheral subunit binding domain and its implications for folding mechanisms.J Mol Biol. 2009 Apr 10;387(4):993-1001. doi: 10.1016/j.jmb.2009.02.014. Epub 2009 Feb 13. J Mol Biol. 2009. PMID: 19217911
-
Atom-by-atom analysis of global downhill protein folding.Nature. 2006 Jul 20;442(7100):317-21. doi: 10.1038/nature04859. Epub 2006 Jun 14. Nature. 2006. PMID: 16799571
-
Downhill versus barrier-limited folding of BBL 2: mechanistic insights from kinetics of folding monitored by independent tryptophan probes.J Mol Biol. 2009 Apr 10;387(4):975-85. doi: 10.1016/j.jmb.2008.12.056. Epub 2008 Dec 30. J Mol Biol. 2009. PMID: 19136014
-
Limited cooperativity in protein folding.Curr Opin Struct Biol. 2016 Feb;36:58-66. doi: 10.1016/j.sbi.2015.12.001. Epub 2016 Feb 2. Curr Opin Struct Biol. 2016. PMID: 26845039 Review.
-
How cooperative are protein folding and unfolding transitions?Protein Sci. 2016 Nov;25(11):1924-1941. doi: 10.1002/pro.3015. Epub 2016 Sep 13. Protein Sci. 2016. PMID: 27522064 Free PMC article. Review.
Cited by
-
Equilibrium unfolding of the PDZ domain of β2-syntrophin.Biophys J. 2012 Jun 20;102(12):2835-44. doi: 10.1016/j.bpj.2012.05.021. Epub 2012 Jun 19. Biophys J. 2012. PMID: 22735534 Free PMC article.
-
Tunable order-disorder continuum in protein-DNA interactions.Nucleic Acids Res. 2018 Sep 28;46(17):8700-8709. doi: 10.1093/nar/gky732. Nucleic Acids Res. 2018. PMID: 30107436 Free PMC article.
-
Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein.Proc Natl Acad Sci U S A. 2013 Oct 1;110(40):E3743-52. doi: 10.1073/pnas.1308381110. Epub 2013 Sep 16. Proc Natl Acad Sci U S A. 2013. PMID: 24043820 Free PMC article.
-
Effects of two solvent conditions on the free energy landscape of the BBL peripheral subunit binding domain.J Phys Chem B. 2012 Jan 12;116(1):646-52. doi: 10.1021/jp209791a. Epub 2011 Dec 13. J Phys Chem B. 2012. PMID: 22126397 Free PMC article.
-
Observation of two families of folding pathways of BBL.Biophys J. 2011 May 18;100(10):2457-65. doi: 10.1016/j.bpj.2011.03.058. Biophys J. 2011. PMID: 21575580 Free PMC article.
References
-
- Dill K. A. (1990) Biochemistry 29, 7133–7155 - PubMed
-
- Fersht A. R. (1998) in Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman W. H. ed) 1st Ed., W. H. Freeman, New York
-
- Poland D. C., Scheraga H. A. (1965) Biopolymers 3, 401–419
-
- Kouza M., Li M. S., O'brien E. P., Jr., Hu C. K., Thirumalai D. (2006) J. Phys. Chem. A. 110, 671–676 - PubMed
-
- Poland D. C., Scheraga H. A. (1965) J. Chem. Phys. 43, 2071–2074 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

