ChIP-Seq identification of weakly conserved heart enhancers
- PMID: 20729851
- PMCID: PMC3138496
- DOI: 10.1038/ng.650
ChIP-Seq identification of weakly conserved heart enhancers
Abstract
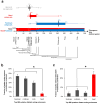
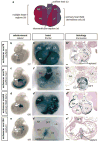
Accurate control of tissue-specific gene expression plays a pivotal role in heart development, but few cardiac transcriptional enhancers have thus far been identified. Extreme noncoding-sequence conservation has successfully predicted enhancers that are active in many tissues but has failed to identify substantial numbers of heart-specific enhancers. Here, we used ChIP-Seq with the enhancer-associated protein p300 from mouse embryonic day 11.5 heart tissue to identify over 3,000 candidate heart enhancers genome wide. Compared to enhancers active in other tissues we studied at this time point, most candidate heart enhancers were less deeply conserved in vertebrate evolution. Nevertheless, transgenic mouse assays of 130 candidate regions revealed that most function reproducibly as enhancers active in the heart, irrespective of their degree of evolutionary constraint. These results provide evidence for a large population of poorly conserved heart enhancers and suggest that the evolutionary conservation of embryonic enhancers can vary depending on tissue type.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Variable evolutionary signatures at the heart of enhancers.Nat Genet. 2010 Sep;42(9):734-5. doi: 10.1038/ng0910-734. Nat Genet. 2010. PMID: 20802475 Free PMC article.
References
-
- Hoffman JI, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am Heart J. 2004;147:425–39. - PubMed
-
- Lloyd-Jones D, et al. Heart disease and stroke statistics--2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:480–6. - PubMed
-
- Pierpont ME, et al. Genetic basis for congenital heart defects: current knowledge: a scientific statement from the American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation. 2007;115:3015–38. - PubMed
-
- Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8. - PubMed
-
- Bentham J, Bhattacharya S. Genetic mechanisms controlling cardiovascular development. Ann N Y Acad Sci. 2008;1123:10–9. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

