A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk
- PMID: 20729854
- PMCID: PMC2930074
- DOI: 10.1038/ng.643
A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk
Abstract
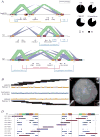
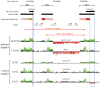
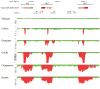
There is a complex relationship between the evolution of segmental duplications and rearrangements associated with human disease. We performed a detailed analysis of one region on chromosome 16p12.1 associated with neurocognitive disease and identified one of the largest structural inconsistencies in the human reference assembly. Various genomic analyses show that all examined humans are homozygously inverted relative to the reference genome for a 1.1-Mb region on 16p12.1. We determined that this assembly discrepancy stems from two common structural configurations with worldwide frequencies of 17.6% (S1) and 82.4% (S2). This polymorphism arose from the rapid integration of segmental duplications, precipitating two local inversions within the human lineage over the last 10 million years. The two human haplotypes differ by 333 kb of additional duplicated sequence present in S2 but not in S1. Notably, we show that the S2 configuration harbors directly oriented duplications, specifically predisposing this chromosome to disease-associated rearrangement.
Conflict of interest statement
E.E.E. is a member of the Scientific Advisory Board Member of Pacific Biosciences. J.A.R. is employee of Signature Genomic Laboratories, LLC. L.G.S. is an employee of, owns shares in and sits on the Members’ Board of Signature Genomic Laboratories, LLC.
Figures

Comment in
-
Large structural polymorphisms predispose genomic sequences to disease-causing rearrangements.Clin Genet. 2011 Feb;79(2):134-5. doi: 10.1111/j.1399-0004.2010.01587.x. Epub 2010 Nov 11. Clin Genet. 2011. PMID: 21070214 No abstract available.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

