Genome instability due to ribonucleotide incorporation into DNA
- PMID: 20729855
- PMCID: PMC2942972
- DOI: 10.1038/nchembio.424
Genome instability due to ribonucleotide incorporation into DNA
Abstract
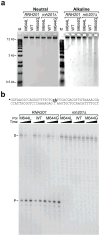
Maintaining the chemical identity of DNA depends on ribonucleotide exclusion by DNA polymerases. However, ribonucleotide exclusion during DNA synthesis in vitro is imperfect. To determine whether ribonucleotides are incorporated during DNA replication in vivo, we substituted leucine or glycine for an active-site methionine in yeast DNA polymerase ϵ (Pol ϵ). Ribonucleotide incorporation in vitro was three-fold lower for M644L and 11-fold higher for M644G Pol ϵ compared to wild-type Pol ϵ. This hierarchy was recapitulated in vivo in yeast strains lacking RNase H2. Moreover, the pol2-M644G rnh201Δ strain progressed more slowly through S phase, had elevated dNTP pools and generated 2-5-base-pair deletions in repetitive sequences at a high rate and in a gene orientation-dependent manner. The data indicate that ribonucleotides are incorporated during replication in vivo, that they are removed by RNase H2-dependent repair and that defective repair results in replicative stress and genome instability via DNA strand misalignment.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Kornberg A, Baker T. DNA replication. Freeman; New York: 1992.
-
- Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem. 1994;140:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

