A polyhedron made of tRNAs
- PMID: 20729899
- PMCID: PMC2928159
- DOI: 10.1038/nchem.733
A polyhedron made of tRNAs
Abstract
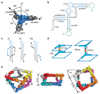
Supramolecular assembly is a powerful strategy used by nature to build nanoscale architectures with predefined sizes and shapes. With synthetic systems, however, numerous challenges remain to be solved before precise control over the synthesis, folding and assembly of rationally designed three-dimensional nano-objects made of RNA can be achieved. Here, using the transfer RNA molecule as a structural building block, we report the design, efficient synthesis and structural characterization of stable, modular three-dimensional particles adopting the polyhedral geometry of a non-uniform square antiprism. The spatial control within the final architecture allows the precise positioning and encapsulation of proteins. This work demonstrates that a remarkable degree of structural control can be achieved with RNA structural motifs for the construction of thermostable three-dimensional nano-architectures that do not rely on helix bundles or tensegrity. RNA three-dimensional particles could potentially be used as carriers or scaffolds in nanomedicine and synthetic biology.
Figures



References
-
- Gesteland RF, Cech TR, Atkins JF. The RNA world. Third Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2005.
-
- Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

