Structural comparison of human mammalian ste20-like kinases
- PMID: 20730082
- PMCID: PMC2920948
- DOI: 10.1371/journal.pone.0011905
Structural comparison of human mammalian ste20-like kinases
Abstract
Background: The serine/threonine mammalian Ste-20 like kinases (MSTs) are key regulators of apoptosis, cellular proliferation as well as polarization. Deregulation of MSTs has been associated with disease progression in prostate and colorectal cancer. The four human MSTs are regulated differently by C-terminal regions flanking the catalytic domains.
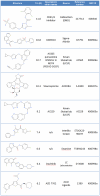
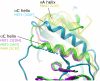
Principal findings: We have determined the crystal structure of kinase domain of MST4 in complex with an ATP-mimetic inhibitor. This is the first structure of an inactive conformation of a member of the MST kinase family. Comparison with active structures of MST3 and MST1 revealed a dimeric association of MST4 suggesting an activation loop exchanged mechanism of MST4 auto-activation. Together with a homology model of MST2 we provide a comparative analysis of the kinase domains for all four members of the human MST family.
Significance: The comparative analysis identified new structural features in the MST ATP binding pocket and has also defined the mechanism for autophosphorylation. Both structural features may be further explored for inhibitors design.
Enhanced version: This article can also be viewed as an enhanced version in which the text of the article is integrated with interactive 3D representations and animated transitions. Please note that a web plugin is required to access this enhanced functionality. Instructions for the installation and use of the web plugin are available in Text S1.
Conflict of interest statement
Figures





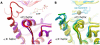

Similar articles
-
Protein phosphatase 2a (PP2A) binds within the oligomerization domain of striatin and regulates the phosphorylation and activation of the mammalian Ste20-Like kinase Mst3.BMC Biochem. 2011 Oct 10;12:54. doi: 10.1186/1471-2091-12-54. BMC Biochem. 2011. PMID: 21985334 Free PMC article.
-
Crystal structure of the PIM2 kinase in complex with an organoruthenium inhibitor.PLoS One. 2009 Oct 20;4(10):e7112. doi: 10.1371/journal.pone.0007112. PLoS One. 2009. PMID: 19841674 Free PMC article.
-
Structures of human MST3 kinase in complex with adenine, ADP and Mn2+.Acta Crystallogr D Biol Crystallogr. 2010 Feb;66(Pt 2):145-54. doi: 10.1107/S0907444909047507. Epub 2010 Jan 22. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20124694
-
SOcK, MiSTs, MASK and STicKs: the GCKIII (germinal centre kinase III) kinases and their heterologous protein-protein interactions.Biochem J. 2013 Aug 15;454(1):13-30. doi: 10.1042/BJ20130219. Biochem J. 2013. PMID: 23889253 Review.
-
Biosignaling of mammalian Ste20-related kinases.Cell Signal. 2008 Jul;20(7):1237-47. doi: 10.1016/j.cellsig.2007.12.019. Epub 2008 Jan 4. Cell Signal. 2008. PMID: 18255267 Review.
Cited by
-
Structural and biochemical insights into the activation mechanisms of germinal center kinase OSR1.J Biol Chem. 2014 Dec 26;289(52):35969-78. doi: 10.1074/jbc.M114.592097. Epub 2014 Nov 11. J Biol Chem. 2014. PMID: 25389294 Free PMC article.
-
Protein Kinase Serine/Threonine Kinase 24 Positively Regulates Interleukin 17-Induced Inflammation by Promoting IKK Complex Activation.Front Immunol. 2018 Apr 30;9:921. doi: 10.3389/fimmu.2018.00921. eCollection 2018. Front Immunol. 2018. PMID: 29760709 Free PMC article.
-
Hippo pathway phylogenetics predicts monoubiquitylation of Salvador and Merlin/Nf2.PLoS One. 2012;7(12):e51599. doi: 10.1371/journal.pone.0051599. Epub 2012 Dec 14. PLoS One. 2012. PMID: 23272121 Free PMC article.
-
miRDRN-miRNA disease regulatory network: a tool for exploring disease and tissue-specific microRNA regulatory networks.PeerJ. 2019 Aug 6;7:e7309. doi: 10.7717/peerj.7309. eCollection 2019. PeerJ. 2019. PMID: 31404401 Free PMC article.
-
Expressing the human proteome for affinity proteomics: optimising expression of soluble protein domains and in vivo biotinylation.N Biotechnol. 2012 Jun 15;29(5):515-25. doi: 10.1016/j.nbt.2011.10.007. Epub 2011 Oct 19. N Biotechnol. 2012. PMID: 22027370 Free PMC article.
References
-
- Eby JJ, Holly SP, van Drogen F, Grishin AV, Peter M, et al. Actin cytoskeleton organization regulated by the PAK family of protein kinases. Curr Biol. 1998;8:967–970. - PubMed
-
- Wu C, Lytvyn V, Thomas DY, Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–30626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

