CTCF prevents the epigenetic drift of EBV latency promoter Qp
- PMID: 20730088
- PMCID: PMC2921154
- DOI: 10.1371/journal.ppat.1001048
CTCF prevents the epigenetic drift of EBV latency promoter Qp
Abstract
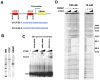
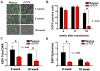
The establishment and maintenance of Epstein-Barr Virus (EBV) latent infection requires distinct viral gene expression programs. These gene expression programs, termed latency types, are determined largely by promoter selection, and controlled through the interplay between cell-type specific transcription factors, chromatin structure, and epigenetic modifications. We used a genome-wide chromatin-immunoprecipitation (ChIP) assay to identify epigenetic modifications that correlate with different latency types. We found that the chromatin insulator protein CTCF binds at several key regulatory nodes in the EBV genome and may compartmentalize epigenetic modifications across the viral genome. Highly enriched CTCF binding sites were identified at the promoter regions upstream of Cp, Wp, EBERs, and Qp. Since Qp is essential for long-term maintenance of viral genomes in type I latency and epithelial cell infections, we focused on the role of CTCF in regulating Qp. Purified CTCF bound approximately 40 bp upstream of the EBNA1 binding sites located at +10 bp relative to the transcriptional initiation site at Qp. Mutagenesis of the CTCF binding site in EBV bacmids resulted in a decrease in the recovery of stable hygromycin-resistant episomes in 293 cells. EBV lacking the Qp CTCF site showed a decrease in Qp transcription initiation and a corresponding increase in Cp and Fp promoter utilization at 8 weeks post-transfection. However, by 16 weeks post-transfection, bacmids lacking CTCF sites had no detectable Qp transcription and showed high levels of histone H3 K9 methylation and CpG DNA methylation at the Qp initiation site. These findings provide direct genetic evidence that CTCF functions as a chromatin insulator that prevents the promiscuous transcription of surrounding genes and blocks the epigenetic silencing of an essential promoter, Qp, during EBV latent infection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Kieff E. Epstein-Barr Virus and its replication.; In: Fields BN, Knipe DM, Howley PM, editors. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. 2 v. (xix, 3091, 3086 p.) p.
-
- Rickinson AB, Kieff E. Epstein-Barr Virus.; In: Fields BN, Knipe DM, Howley PM, editors. Philadelphia: Wolters Kluwer Health/Lippincott Williams & Wilkins; 2007. 2 v. (xix, 3091, 3086 p.) p.
-
- Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. - PubMed
-
- Babcock GJ, Hochberg D, Thorley-Lawson AD. The expression pattern of Epstein-Barr virus latent genes in vivo is dependent upon the differentiation stage of the infected B cell. Immunity. 2000;13:497–506. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

