Conserved and distinct modes of CREB/ATF transcription factor regulation by PP2A/B56gamma and genotoxic stress
- PMID: 20730097
- PMCID: PMC2921338
- DOI: 10.1371/journal.pone.0012173
Conserved and distinct modes of CREB/ATF transcription factor regulation by PP2A/B56gamma and genotoxic stress
Abstract
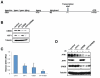
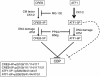
Activating transcription factor 1 (ATF1) and the closely related proteins CREB (cyclic AMP resonse element binding protein) and CREM (cyclic AMP response element modulator) constitute a subfamily of bZIP transcription factors that play critical roles in the regulation of cellular growth, metabolism, and survival. Previous studies demonstrated that CREB is phosphorylated on a cluster of conserved Ser residues, including Ser-111 and Ser-121, in response to DNA damage through the coordinated actions of the ataxia-telangiectasia-mutated (ATM) protein kinase and casein kinases 1 and 2 (CK1/2). Here, we show that DNA damage-induced phosphorylation by ATM is a general feature of CREB and ATF1. ATF1 harbors a conserved ATM/CK cluster that is constitutively and stoichiometrically phosphorylated by CK1 and CK2 in asynchronously growing cells. Exposure to DNA damage further induced ATF1 phosphorylation on Ser-51 by ATM in a manner that required prior phosphorylation of the upstream CK residues. Hyperphosphorylated ATF1 showed a 4-fold reduced affinity for CREB-binding protein. We further show that PP2A, in conjunction with its targeting subunit B56gamma, antagonized ATM and CK1/2-dependent phosphorylation of CREB and ATF1 in cellulo. Finally, we show that CK sites in CREB are phosphorylated during cellular growth and that phosphorylation of these residues reduces the threshold of DNA damage required for ATM-dependent phosphorylation of the inhibitory Ser-121 residue. These studies define overlapping and distinct modes of CREB and ATF1 regulation by phosphorylation that may ensure concerted changes in gene expression mediated by these factors.
Conflict of interest statement
Figures




Similar articles
-
Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism.Nucleic Acids Res. 2016 Nov 16;44(20):9667-9680. doi: 10.1093/nar/gkw643. Epub 2016 Jul 18. Nucleic Acids Res. 2016. PMID: 27431323 Free PMC article.
-
Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage.J Biol Chem. 2007 Mar 2;282(9):6283-91. doi: 10.1074/jbc.M610674200. Epub 2007 Jan 5. J Biol Chem. 2007. PMID: 17209043
-
DNA replication stress-induced phosphorylation of cyclic AMP response element-binding protein mediated by ATM.J Biol Chem. 2006 Jan 20;281(3):1692-7. doi: 10.1074/jbc.M509577200. Epub 2005 Nov 17. J Biol Chem. 2006. PMID: 16293623
-
Expression, molecular mechanisms and therapeutic potentials of ATF1 in cancers.Life Sci. 2025 Jan 1;360:123256. doi: 10.1016/j.lfs.2024.123256. Epub 2024 Nov 21. Life Sci. 2025. PMID: 39580140 Review.
-
Molecular recognition by the KIX domain and its role in gene regulation.Nucleic Acids Res. 2014 Feb;42(4):2112-25. doi: 10.1093/nar/gkt1147. Epub 2013 Nov 18. Nucleic Acids Res. 2014. PMID: 24253305 Free PMC article. Review.
Cited by
-
Structural Basis for Graded Inhibition of CREB:DNA Interactions by Multisite Phosphorylation.Biochemistry. 2018 Dec 26;57(51):6964-6972. doi: 10.1021/acs.biochem.8b01092. Epub 2018 Dec 13. Biochemistry. 2018. PMID: 30507144 Free PMC article.
-
What turns CREB on? And off? And why does it matter?Cell Mol Life Sci. 2020 Oct;77(20):4049-4067. doi: 10.1007/s00018-020-03525-8. Epub 2020 Apr 28. Cell Mol Life Sci. 2020. PMID: 32347317 Free PMC article. Review.
-
ATF1 modulates the heat shock response by regulating the stress-inducible heat shock factor 1 transcription complex.Mol Cell Biol. 2015 Jan;35(1):11-25. doi: 10.1128/MCB.00754-14. Epub 2014 Oct 13. Mol Cell Biol. 2015. PMID: 25312646 Free PMC article.
-
Novel Ser/Thr protein phosphatases in cell death regulation.Physiology (Bethesda). 2012 Feb;27(1):43-52. doi: 10.1152/physiol.00034.2011. Physiology (Bethesda). 2012. PMID: 22311969 Free PMC article. Review.
-
Tunable regulation of CREB DNA binding activity couples genotoxic stress response and metabolism.Nucleic Acids Res. 2016 Nov 16;44(20):9667-9680. doi: 10.1093/nar/gkw643. Epub 2016 Jul 18. Nucleic Acids Res. 2016. PMID: 27431323 Free PMC article.
References
-
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. - PubMed
-
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. - PubMed
-
- Montminy M, Koo SH, Zhang X. The CREB family: key regulators of hepatic metabolism. Ann Endocrinol (Paris) 2004;65:73–75. - PubMed
-
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

