Live cell imaging and systems biology
- PMID: 20730797
- PMCID: PMC2992103
- DOI: 10.1002/wsbm.108
Live cell imaging and systems biology
Abstract
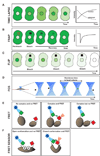
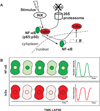
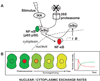
Much of the experimental data used to construct mathematical models of molecular networks are derived from in vitro measurements. However, there is increasing evidence that in vitro measurements fail to capture both the complexity and the individuality found in single, living cells. These limitations can be overcome by live cell microscopy which is evolving to enable in vivo biochemistry. Here, we survey the current capabilities of live cell microscopy and illustrate how a number of different imaging approaches could be applied to analyze a specific molecular network. We argue that incorporation of such quantitative live-cell imaging methods is critical for the progress of systems biology.
Copyright © 2010 John Wiley & Sons, Inc.
Figures

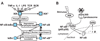
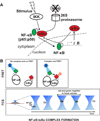

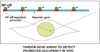
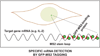
References
-
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. - PubMed
-
- Nelson DE, Ihekwaba AE, Elliott M, Johnson JR, Gibney CA, Foreman BE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. - PubMed
-
- Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545–550. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

