Understanding the effect of magnesium ion concentration on the catalytic activity of ribonuclease H through computation: does a third metal binding site modulate endonuclease catalysis?
- PMID: 20731347
- PMCID: PMC2989666
- DOI: 10.1021/ja102933y
Understanding the effect of magnesium ion concentration on the catalytic activity of ribonuclease H through computation: does a third metal binding site modulate endonuclease catalysis?
Abstract
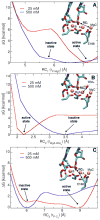
Ribonuclease H (RNase H) belongs to the nucleotidyl-transferase superfamily and hydrolyzes the phosphodiester linkage on the RNA strand of a DNA/RNA hybrid duplex. Due to its activity in HIV reverse transcription, it represents a promising target for anti-HIV drug design. While crystallographic data have located two ions in the catalytic site, there is ongoing debate concerning just how many metal ions bound at the active site are optimal for catalysis. Indeed, experiments have shown a dependency of the catalytic activity on the Mg(2+) concentration. Moreover, in RNase H, the glutamate residue E188 has been shown to be essential for full enzymatic activation, regardless of the Mg(2+) concentration. The catalytic center is known to contain two Mg(2+) ions, and E188 is not one of the primary metal ligands. Herein, classical molecular dynamics (MD) simulations are employed to study the metal-ligand coordination in RNase H at different concentration of Mg(2+). Importantly, the presence of a third Mg(2+) ion, bound to the second-shell ligand E188, is a persistent feature of the MD simulations. Free energy calculations have identified two distinct conformations, depending on the concentration of Mg(2+). At standard concentration, a third Mg(2+) is found in the catalytic pocket, but it does not perturb the optimal RNase H active conformation. However, at higher concentration, the third Mg(2+) ion heavily perturbs the nucleophilic water and thereby influences the catalytic efficiency of RNase H. In addition, the E188A mutant shows no ability to engage additional Mg(2+) ions near the catalytic pocket. This finding likely explains the decrease in catalytic activity of E188A and also supports the key role of E188 in localizing the third Mg(2+) ion at the active site. Glutamate residues are commonly found surrounding the metal center in the endonuclease family, which suggests that this structural motif may be an important feature to enhance catalytic activity. The present MD calculations support the hypothesis that RNase H can accommodate three divalent metal ions in its catalytic pocket and provide an in-depth understanding of their dynamic role for catalysis.
Figures





Similar articles
-
Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism.J Am Chem Soc. 2008 Aug 20;130(33):10955-62. doi: 10.1021/ja8005786. Epub 2008 Jul 29. J Am Chem Soc. 2008. PMID: 18662000 Free PMC article.
-
Co-crystal of Escherichia coli RNase HI with Mn2+ ions reveals two divalent metals bound in the active site.J Biol Chem. 2001 Mar 9;276(10):7266-71. doi: 10.1074/jbc.M009626200. Epub 2000 Nov 16. J Biol Chem. 2001. PMID: 11083878
-
Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis.Cell. 2005 Jul 1;121(7):1005-16. doi: 10.1016/j.cell.2005.04.024. Cell. 2005. PMID: 15989951
-
Catalytic metal ions and enzymatic processing of DNA and RNA.Acc Chem Res. 2015 Feb 17;48(2):220-8. doi: 10.1021/ar500314j. Epub 2015 Jan 15. Acc Chem Res. 2015. PMID: 25590654 Review.
-
Identification of catalytic metal ion ligands in ribozymes.Methods. 2009 Oct;49(2):148-66. doi: 10.1016/j.ymeth.2009.07.005. Epub 2009 Aug 3. Methods. 2009. PMID: 19651216 Free PMC article. Review.
Cited by
-
Calcium inhibition of ribonuclease H1 two-metal ion catalysis.J Am Chem Soc. 2014 Feb 26;136(8):3137-44. doi: 10.1021/ja411408x. Epub 2014 Feb 18. J Am Chem Soc. 2014. PMID: 24499076 Free PMC article.
-
Optimizing Messenger RNA Analysis Using Ultra-Wide Pore Size Exclusion Chromatography Columns.Int J Mol Sci. 2024 Jun 6;25(11):6254. doi: 10.3390/ijms25116254. Int J Mol Sci. 2024. PMID: 38892442 Free PMC article.
-
Evidence for a dual functional role of a conserved histidine in RNA·DNA heteroduplex cleavage by human RNase H1.FEBS J. 2012 Dec;279(24):4492-500. doi: 10.1111/febs.12035. Epub 2012 Nov 9. FEBS J. 2012. PMID: 23078533 Free PMC article.
-
Impact of hierarchical water dipole orderings on the dynamics of aqueous salt solutions.Nat Commun. 2023 Aug 7;14(1):4616. doi: 10.1038/s41467-023-40278-x. Nat Commun. 2023. PMID: 37550299 Free PMC article.
-
Structural basis for Ca2+-dependent catalysis of a cutinase-like enzyme and its engineering: application to enzymatic PET depolymerization.Biophys Physicobiol. 2021 Jun 30;18:168-176. doi: 10.2142/biophysico.bppb-v18.018. eCollection 2021. Biophys Physicobiol. 2021. PMID: 34386313 Free PMC article.
References
-
- Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M, Matsuzaki T, Morikawa K. Nature. 1990;347:306–309. - PubMed
-
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Cell. 2005;121:1005–1016. - PubMed
-
- Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W. Molecular Cell. 2007;28:264–276. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

