Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity
- PMID: 20731629
- PMCID: PMC2995810
- DOI: 10.1111/j.1369-1600.2010.00248.x
Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity
Abstract
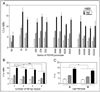
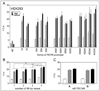
A polymorphic 68-bp tandem repeat has been identified within the promoter of the human prodynorphin (PDYN) gene. We found that this 68-bp repeat in the PDYN promoter occurs naturally up to five times. We studied the effect of the number of 68-bp repeats, and of a SNP (rs61761346) found within the repeat on PDYN gene promoter activity. Thirteen promoter forms, different naturally occurring combinations of repeats and the internal SNP, were cloned upstream of the luciferase reporter gene, transfected into human SK-N-SH, H69, or HEK293 cells. Cells were then stimulated with TPA or caffeine. We found cell-specific effects of the number of 68-bp repeats on the transcriptional activity of the PDYN promoter. In SK-N-SH and H69 cells, three or four repeats led to lower expression of luciferase than did one or two repeats. The opposite effect was found in HEK293 cells. The SNP also had an effect on PDYN gene expression in both SK-N-SH and H69 cells; promoter forms with the A allele had significantly higher expression than promoter forms with the G allele. These results further our understanding of the complex transcriptional regulation of the PDYN gene promoter.
© 2010 The Authors, Addiction Biology © 2010 Society for the Study of Addiction.
Figures




Similar articles
-
Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence.Addict Biol. 2007 Sep;12(3-4):496-502. doi: 10.1111/j.1369-1600.2007.00069.x. Epub 2007 Jun 8. Addict Biol. 2007. PMID: 17559549
-
Potentially functional polymorphism in the promoter region of prodynorphin gene may be associated with protection against cocaine dependence or abuse.Am J Med Genet. 2002 May 8;114(4):429-35. doi: 10.1002/ajmg.10362. Am J Med Genet. 2002. PMID: 11992566 Free PMC article.
-
Prodynorphin promoter SNP associated with alcohol dependence forms noncanonical AP-1 binding site that may influence gene expression in human brain.Brain Res. 2011 Apr 18;1385:18-25. doi: 10.1016/j.brainres.2011.02.042. Epub 2011 Feb 19. Brain Res. 2011. PMID: 21338584
-
Association between VNTR polymorphism in promoter region of prodynorphin (PDYN) gene and heroin dependence.Psychiatry Res. 2014 Nov 30;219(3):690-2. doi: 10.1016/j.psychres.2014.06.048. Epub 2014 Jul 5. Psychiatry Res. 2014. PMID: 25048760
-
The alpha and eta isoforms of protein kinase C stimulate transcription of human involucrin gene.J Invest Dermatol. 1998 Mar;110(3):218-23. doi: 10.1046/j.1523-1747.1998.00110.x. J Invest Dermatol. 1998. PMID: 9506439 Review.
Cited by
-
Polymorphisms of the kappa opioid receptor and prodynorphin genes: HIV risk and HIV natural history.J Acquir Immune Defic Syndr. 2013 May 1;63(1):17-26. doi: 10.1097/QAI.0b013e318285cd0c. J Acquir Immune Defic Syndr. 2013. PMID: 23392455 Free PMC article.
-
Tissue-specific DNA methylation of the human prodynorphin gene in post-mortem brain tissues and PBMCs.Pharmacogenet Genomics. 2011 Apr;21(4):185-96. doi: 10.1097/FPC.0b013e32833eecbc. Pharmacogenet Genomics. 2011. PMID: 20808262 Free PMC article.
-
κ-opioid receptor/dynorphin system: genetic and pharmacotherapeutic implications for addiction.Trends Neurosci. 2012 Oct;35(10):587-96. doi: 10.1016/j.tins.2012.05.005. Epub 2012 Jun 16. Trends Neurosci. 2012. PMID: 22709632 Free PMC article. Review.
-
Epigenetic and Transcriptional Control of the Opioid Prodynorphine Gene: In-Depth Analysis in the Human Brain.Molecules. 2021 Jun 7;26(11):3458. doi: 10.3390/molecules26113458. Molecules. 2021. PMID: 34200173 Free PMC article. Review.
-
Genetic association analyses of PDYN polymorphisms with heroin and cocaine addiction.Genes Brain Behav. 2012 Jun;11(4):415-23. doi: 10.1111/j.1601-183X.2012.00785.x. Epub 2012 Apr 11. Genes Brain Behav. 2012. PMID: 22443215 Free PMC article.
References
-
- Babbitt CC, Silverman JS, Haygood R, Reininga JM, Rockman MV, Wray GA. Multiple functional variants in cis modulate PDYN expression. Mol Biol Evol. 2010;27:465–479. - PubMed
-
- Bovo G, Diani E, Bisulli F, Di Bonaventura C, Striano P, Gambardella A, Ferlazzo E, Egeo G, Mecarelli O, Elia M, Bianchi A, Bortoluzzi S, Vettori A, Aguglia U, Binelli S, De Falco A, Coppola G, Gobbi G, Sofia V, Striano S, Tinuper P, Giallonardo AT, Michelucci R, Nobile C. Analysis of LGI1 promoter sequence, PDYN and GABBR1 polymorphisms in sporadic and familial lateral temporal lobe epilepsy. Neurosci. Lett. 2008;436:23–26. - PubMed
-
- Campos D, Jimenez-Diaz L, Carrion AM. Ca(2+)-dependent prodynorphin transcriptional derepression in neuroblastoma cells is exerted through DREAM protein activity in a kinase-independent manner. Mol. Cell Neurosci. 2003;22:135–145. - PubMed
-
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

