Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses
- PMID: 20732878
- PMCID: PMC2935864
- DOI: 10.1093/jxb/erq196
Subgroup 4 R2R3-MYBs in conifer trees: gene family expansion and contribution to the isoprenoid- and flavonoid-oriented responses
Abstract
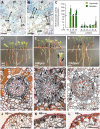
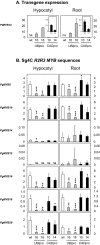
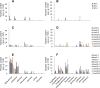
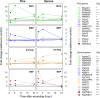
Transcription factors play a fundamental role in plants by orchestrating temporal and spatial gene expression in response to environmental stimuli. Several R2R3-MYB genes of the Arabidopsis subgroup 4 (Sg4) share a C-terminal EAR motif signature recently linked to stress response in angiosperm plants. It is reported here that nearly all Sg4 MYB genes in the conifer trees Picea glauca (white spruce) and Pinus taeda (loblolly pine) form a monophyletic clade (Sg4C) that expanded following the split of gymnosperm and angiosperm lineages. Deeper sequencing in P. glauca identified 10 distinct Sg4C sequences, indicating over-representation of Sg4 sequences compared with angiosperms such as Arabidopsis, Oryza, Vitis, and Populus. The Sg4C MYBs share the EAR motif core. Many of them had stress-responsive transcript profiles after wounding, jasmonic acid (JA) treatment, or exposure to cold in P. glauca and P. taeda, with MYB14 transcripts accumulating most strongly and rapidly. Functional characterization was initiated by expressing the P. taeda MYB14 (PtMYB14) gene in transgenic P. glauca plantlets with a tissue-preferential promoter (cinnamyl alcohol dehydrogenase) and a ubiquitous gene promoter (ubiquitin). Histological, metabolite, and transcript (microarray and targeted quantitative real-time PCR) analyses of PtMYB14 transgenics, coupled with mechanical wounding and JA application experiments on wild-type plantlets, allowed identification of PtMYB14 as a putative regulator of an isoprenoid-oriented response that leads to the accumulation of sesquiterpene in conifers. Data further suggested that PtMYB14 may contribute to a broad defence response implicating flavonoids. This study also addresses the potential involvement of closely related Sg4C sequences in stress responses and plant evolution.
Figures


References
-
- Agarwal M, Hao Y, Kapoor A, Dong C-H, Fujii H, Zheng X, Zhu J- K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. - PubMed
-
- Agrawal AA. Macroevolution of plant defense strategies. Trends in Ecology & Evolution. 2007;22:103–109. - PubMed
-
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings/International Conference on Intelligent Systems for Molecular Biology. 1994;2:28–36. - PubMed
-
- Bedon F, Levasseur C, Grima-Pettenati J, Séguin A, MacKay J. Sequence analysis and functional characterization of the promoter of the Picea glauca cinnamyl alcohol dehydrogenase gene in transgenic white spruce plants. Plant Cell Reports. 2009;28:787–800. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

