Rap1b facilitates NK cell functions via IQGAP1-mediated signalosomes
- PMID: 20733035
- PMCID: PMC2931159
- DOI: 10.1084/jem.20100040
Rap1b facilitates NK cell functions via IQGAP1-mediated signalosomes
Abstract
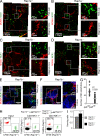
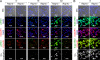
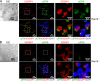
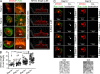
Rap1 GTPases control immune synapse formation and signaling in lymphocytes. However, the precise molecular mechanism by which Rap1 regulates natural killer (NK) cell activation is not known. Using Rap1a or Rap1b knockout mice, we identify Rap1b as the major isoform in NK cells. Its absence significantly impaired LFA1 polarization, spreading, and microtubule organizing center (MTOC) formation in NK cells. Neither Rap1 isoform was essential for NK cytotoxicity. However, absence of Rap1b impaired NKG2D, Ly49D, and NCR1-mediated cytokine and chemokine production. Upon activation, Rap1b colocalized with the scaffolding protein IQGAP1. This interaction facilitated sequential phosphorylation of B-Raf, C-Raf, and ERK1/2 and helped IQGAP1 to form a large signalosome in the perinuclear region. These results reveal a previously unrecognized role for Rap1b in NK cell signaling and effector functions.
Figures




References
-
- Barber D.F., Faure M., Long E.O. 2004. LFA-1 contributes an early signal for NK cell cytotoxicity. J. Immunol. 173:3653–3659 - PubMed
-
- Berenson L.S., Yang J., Sleckman B.P., Murphy T.L., Murphy K.M. 2006. Selective requirement of p38alpha MAPK in cytokine-dependent, but not antigen receptor-dependent, Th1 responses. J. Immunol. 176:4616–4621 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

