A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA
- PMID: 20733054
- PMCID: PMC2928013
- DOI: 10.1083/jcb.201002124
A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA
Abstract
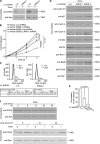
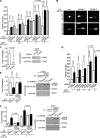
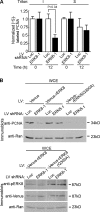
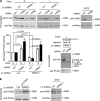
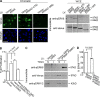
Proliferating cell nuclear antigen (PCNA) acts as a scaffold, coordinator, and stimulator of numerous processes required for faithful transmission of genetic information. Maintaining PCNA levels above a critical threshold is essential, but little is known about PCNA protein turnover. We now show that ERK8 (extracellular signal-regulated kinase 8) is required for PCNA protein stability. ERK8 contains a conserved PCNA-interacting protein (PIP) box. Chromatin-bound ERK8 (ERK8(CHROMATIN)) interacts via this motif with PCNA(CHROMATIN), which acts as a platform for numerous proteins involved in DNA metabolism. Silencing ERK8 decreases PCNA levels and increases DNA damage. Ectopic expression of PCNA blocks DNA damage induced by ERK8 loss. ERK8 prevents HDM2-mediated PCNA destruction by inhibiting the association of PCNA with HDM2. This regulation is physiologically relevant as ERK8 activity is inhibited in transformed mammary cells. Our results reveal an unanticipated mechanism to control PCNA levels in normal cycling mammary epithelial cells and implicate ERK8 in the regulation of genomic stability.
Figures


Comment in
-
HDM2 ERKs PCNA.J Cell Biol. 2010 Aug 23;190(4):487-9. doi: 10.1083/jcb.201007096. J Cell Biol. 2010. PMID: 20733049 Free PMC article.
Similar articles
-
HDM2 ERKs PCNA.J Cell Biol. 2010 Aug 23;190(4):487-9. doi: 10.1083/jcb.201007096. J Cell Biol. 2010. PMID: 20733049 Free PMC article.
-
Regulation of PCNA-protein interactions for genome stability.Nat Rev Mol Cell Biol. 2013 May;14(5):269-82. doi: 10.1038/nrm3562. Epub 2013 Apr 18. Nat Rev Mol Cell Biol. 2013. PMID: 23594953 Review.
-
PCNA-dependent ubiquitination of Cdt1 and p21 in mammalian cells.Methods Mol Biol. 2014;1170:367-82. doi: 10.1007/978-1-4939-0888-2_19. Methods Mol Biol. 2014. PMID: 24906324
-
PCNA antagonizes cohesin-dependent roles in genomic stability.PLoS One. 2020 Oct 19;15(10):e0235103. doi: 10.1371/journal.pone.0235103. eCollection 2020. PLoS One. 2020. PMID: 33075068 Free PMC article.
-
PCNA cycling dynamics during DNA replication and repair in mammals.Trends Genet. 2024 Jun;40(6):526-539. doi: 10.1016/j.tig.2024.02.006. Epub 2024 Mar 13. Trends Genet. 2024. PMID: 38485608 Review.
Cited by
-
ERKing Trypanosoma: PCNA phosphorylation as novel target.Cell Cycle. 2016 Dec;15(23):3167-3168. doi: 10.1080/15384101.2016.1232081. Epub 2016 Sep 16. Cell Cycle. 2016. PMID: 27635479 Free PMC article. No abstract available.
-
Extracellular signal-regulated kinase 8 (ERK8) controls estrogen-related receptor α (ERRα) cellular localization and inhibits its transcriptional activity.J Biol Chem. 2011 Mar 11;286(10):8507-8522. doi: 10.1074/jbc.M110.179523. Epub 2010 Dec 28. J Biol Chem. 2011. PMID: 21190936 Free PMC article.
-
A FRET-Based Assay for the Identification of PCNA Inhibitors.Int J Mol Sci. 2023 Jul 24;24(14):11858. doi: 10.3390/ijms241411858. Int J Mol Sci. 2023. PMID: 37511614 Free PMC article.
-
MAPK15 protects from oxidative stress-dependent cellular senescence by inducing the mitophagic process.Aging Cell. 2022 Jul;21(7):e13620. doi: 10.1111/acel.13620. Epub 2022 Jun 1. Aging Cell. 2022. PMID: 35642724 Free PMC article.
-
ERα-Mediated Nuclear Sequestration of RSK2 Is Required for ER+ Breast Cancer Tumorigenesis.Cancer Res. 2018 Apr 15;78(8):2014-2025. doi: 10.1158/0008-5472.CAN-17-2063. Epub 2018 Jan 19. Cancer Res. 2018. PMID: 29351904 Free PMC article.
References
-
- Banks D., Wu M., Higa L.A., Gavrilova N., Quan J., Ye T., Kobayashi R., Sun H., Zhang H. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle. 5:1719–1729 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

