Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499
- PMID: 20733065
- PMCID: PMC3057038
- DOI: 10.1161/CIRCGENETICS.109.934281
Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499
Abstract
Background: MicroRNAs (miRNAs) are a newly discovered endogenous class of small, noncoding RNAs that play important posttranscriptional regulatory roles by targeting messenger RNAs for cleavage or translational repression. Human embryonic stem cells are known to express miRNAs that are often undetectable in adult organs, and a growing body of evidence has implicated miRNAs as important arbiters of heart development and disease.
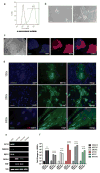
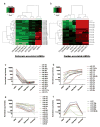
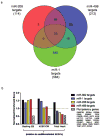
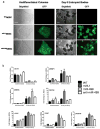
Methods and results: To better understand the transition between the human embryonic and cardiac "miRNA-omes," we report here the first miRNA profiling study of cardiomyocytes derived from human embryonic stem cells. Analyzing 711 unique miRNAs, we have identified several interesting miRNAs, including miR-1, -133, and -208, that have been previously reported to be involved in cardiac development and disease and that show surprising patterns of expression across our samples. We also identified novel miRNAs, such as miR-499, that are strongly associated with cardiac differentiation and that share many predicted targets with miR-208. Overexpression of miR-499 and -1 resulted in upregulation of important cardiac myosin heavy-chain genes in embryoid bodies; miR-499 overexpression also caused upregulation of the cardiac transcription factor MEF2C.
Conclusions: Taken together, our data give significant insight into the regulatory networks that govern human embryonic stem cell differentiation and highlight the ability of miRNAs to perturb, and even control, the genes that are involved in cardiac specification of human embryonic stem cells.
Conflict of interest statement
Figures
Comment in
-
Letter by van Mil et al regarding, "Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499".Circ Cardiovasc Genet. 2011 Feb;4(1):e3; author reply e4. doi: 10.1161/CIRCGENETICS.110.958595. Circ Cardiovasc Genet. 2011. PMID: 21325154 No abstract available.
References
-
- van Laake LW, Passier R, Doevendans PA, Mummery CL. Human embryonic stem cell-derived cardiomyocytes and cardiac repair in rodents. Circulation Research. 2008;102:1008–10. - PubMed
-
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill EA, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nature Biotechnology. 2007;25:1015–24. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

