Structure-based modeling of the functional HIV-1 intasome and its inhibition
- PMID: 20733078
- PMCID: PMC2936642
- DOI: 10.1073/pnas.1002346107
Structure-based modeling of the functional HIV-1 intasome and its inhibition
Abstract
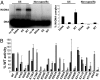
The intasome is the basic recombination unit of retroviral integration, comprising the integrase protein and the ends of the viral DNA made by reverse transcription. Clinical inhibitors preferentially target the DNA-bound form of integrase as compared with the free protein, highlighting the critical requirement for detailed understanding of HIV-1 intasome structure and function. Although previous biochemical studies identified integrase residues that contact the DNA, structural details of protein-protein and protein-DNA interactions within the functional intasome were lacking. The recent crystal structure of the prototype foamy virus (PFV) integrase-viral DNA complex revealed numerous details of this related integration machine. Structures of drug-bound PFV intasomes moreover elucidated the mechanism of inhibitor action. Herein we present a model for the HIV-1 intasome assembled using the PFV structure as template. Our results pinpoint previously identified protein-DNA contacts within the quaternary structure and reveal hitherto unknown roles for Arg20 and Lys266 in DNA binding and integrase function. Models for clinical inhibitors bound at the HIV-1 integrase active site were also constructed and compared with previous studies. Our findings highlight the structural basis for HIV-1 integration and define the mechanism of its inhibition, which should help in formulating new drugs to inhibit viruses resistant to first-in-class compounds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

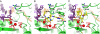
Similar articles
-
Modeling the HIV-1 Intasome: A Prototype View of the Target of Integrase Inhibitors.Viruses. 2010 Dec;2(12):2777-81. doi: 10.3390/v2122777. Epub 2010 Dec 21. Viruses. 2010. PMID: 21994639 Free PMC article.
-
HIV-1 Intasomes Assembled with Excess Integrase C-Terminal Domain Protein Facilitate Structural Studies by Cryo-EM and Reveal the Role of the Integrase C-Terminal Tail in HIV-1 Integration.Viruses. 2024 Jul 20;16(7):1166. doi: 10.3390/v16071166. Viruses. 2024. PMID: 39066328 Free PMC article.
-
Outer domains of integrase within retroviral intasomes are dispensible for catalysis of DNA integration.Protein Sci. 2016 Feb;25(2):472-8. doi: 10.1002/pro.2837. Epub 2015 Nov 25. Protein Sci. 2016. PMID: 26537415 Free PMC article.
-
Retroviral integrase protein and intasome nucleoprotein complex structures.World J Biol Chem. 2017 Feb 26;8(1):32-44. doi: 10.4331/wjbc.v8.i1.32. World J Biol Chem. 2017. PMID: 28289517 Free PMC article. Review.
-
Structural Biology of HIV Integrase Strand Transfer Inhibitors.Trends Pharmacol Sci. 2020 Sep;41(9):611-626. doi: 10.1016/j.tips.2020.06.003. Epub 2020 Jul 3. Trends Pharmacol Sci. 2020. PMID: 32624197 Free PMC article. Review.
Cited by
-
Conversion of raltegravir carrying a 1,3,4-oxadiazole ring to a hydrolysis product upon pH changes decreases its antiviral activity.PNAS Nexus. 2023 Dec 18;3(1):pgad446. doi: 10.1093/pnasnexus/pgad446. eCollection 2024 Jan. PNAS Nexus. 2023. PMID: 38170115 Free PMC article.
-
Effect of HIV-1 integrase resistance mutations when introduced into SIVmac239 on susceptibility to integrase strand transfer inhibitors.J Virol. 2014 Sep 1;88(17):9683-92. doi: 10.1128/JVI.00947-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920794 Free PMC article.
-
Design of HIV-1 integrase inhibitors targeting the catalytic domain as well as its interaction with LEDGF/p75: a scaffold hopping approach using salicylate and catechol groups.Bioorg Med Chem. 2011 Aug 15;19(16):4935-52. doi: 10.1016/j.bmc.2011.06.058. Epub 2011 Jun 26. Bioorg Med Chem. 2011. PMID: 21778063 Free PMC article.
-
The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration.Biochemistry. 2011 Nov 15;50(45):9788-96. doi: 10.1021/bi201247f. Epub 2011 Oct 19. Biochemistry. 2011. PMID: 21992419 Free PMC article.
-
Interaction of HIV-1 integrase with polypyrimidine tract binding protein and associated splicing factor (PSF) and its impact on HIV-1 replication.Retrovirology. 2019 Apr 29;16(1):12. doi: 10.1186/s12977-019-0474-1. Retrovirology. 2019. PMID: 31036027 Free PMC article.
References
-
- Engelman A. Reverse transcription and integration. In: Kurth R, Bannert N, editors. Retroviruses: Molecular Biology, Genomics and Pathogenesis. Norfolk, United Kingdom: Caister Academic Press; 2010. pp. 129–159.
-
- Dyda F, et al. Crystal structure of the catalytic domain of HIV-1 integrase: Similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. - PubMed
-
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. - PubMed
-
- Farnet CM, Bushman FD. HIV-1 cDNA integration: Requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

