Conformational dependence of 13C shielding and coupling constants for methionine methyl groups
- PMID: 20734113
- PMCID: PMC5598763
- DOI: 10.1007/s10858-010-9436-6
Conformational dependence of 13C shielding and coupling constants for methionine methyl groups
Abstract
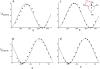
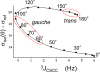
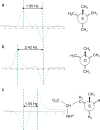
Methionine residues fulfill a broad range of roles in protein function related to conformational plasticity, ligand binding, and sensing/mediating the effects of oxidative stress. A high degree of internal mobility, intrinsic detection sensitivity of the methyl group, and low copy number have made methionine labeling a popular approach for NMR investigation of selectively labeled protein macromolecules. However, selective labeling approaches are subject to more limited information content. In order to optimize the information available from such studies, we have performed DFT calculations on model systems to evaluate the conformational dependence of (3)J (CSCC), (3)J (CSCH), and the isotropic shielding, sigma(iso). Results have been compared with experimental data reported in the literature, as well as data obtained on [methyl-(13)C]methionine and on model compounds. These studies indicate that relative to oxygen, the presence of the sulfur atom in the coupling pathway results in a significantly smaller coupling constant, (3)J (CSCC)/(3)J (COCC) approximately 0.7. It is further demonstrated that the (3)J (CSCH) coupling constant depends primarily on the subtended CSCH dihedral angle, and secondarily on the CSCC dihedral angle. Comparison of theoretical shielding calculations with the experimental shift range of the methyl group for methionine residues in proteins supports the conclusion that the intra-residue conformationally-dependent shift perturbation is the dominant determinant of delta(13)Cepsilon. Analysis of calmodulin data based on these calculations indicates that several residues adopt non-standard rotamers characterized by very large approximately 100 degrees chi(3) values. The utility of the delta(13)Cepsilon as a basis for estimating the gauche/trans ratio for chi(3) is evaluated, and physical and technical factors that limit the accuracy of both the NMR and crystallographic analyses are discussed.
Figures


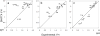


References
-
- Anbanandam A, Bieber Urbauer RJ, Bartlett RK, Smallwood HS, Squier TC, Urbauer JL. Mediating molecular recognition by methionine oxidation: conformational switching by oxidation of methionine in the carboxyl-terminal domain of calmodulin. Biochemistry. 2005;44:9486–9496. - PubMed
-
- Babu YS, Bugg CE, Cook WJ. Structure of calmodulin refined at 2.2 A resolution. J Mol Biol. 1988;204:191–204. - PubMed
-
- Barfield M, Marshall JL, Canada ED. Nuclear Spin-Spin Coupling Via Nonbonded Interactions .2. Gamma-Substituent Effects for Vicinal Coupling-Constants Involving C-13. Journal of the American Chemical Society. 1980;102:7–12.
-
- Batchelor JG, Feeney J, Roberts GCK. C-13 Nmr Protonation Shifts of Amines, Carboxylic-Acids and Amino-Acids. Journal of Magnetic Resonance. 1975;20:19–38.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

