Caspases: evolutionary aspects of their functions in vertebrates
- PMID: 20735596
- PMCID: PMC2779465
- DOI: 10.1111/j.1095-8649.2009.02184.x
Caspases: evolutionary aspects of their functions in vertebrates
Abstract
Caspases (cysteine-dependent aspartyl-specific protease) belong to a family of cysteine proteases that mediate proteolytic events indispensable for biological phenomena such as cell death and inflammation. The first caspase was identified as an executioner of apoptotic cell death in the worm Caenorhabditis elegans. Additionally, a large number of caspases have been identified in various animals from sponges to vertebrates. Caspases are thought to play a pivotal role in apoptosis as an evolutionarily conserved function; however, the number of caspases that can be identified is distinct for each species. This indicates that species-specific functions or diversification of physiological roles has been cultivated through caspase evolution. Furthermore, recent studies suggest that caspases are also involved in inflammation and cellular differentiation in mammals. This review highlights vertebrate caspases in their universal and divergent functions and provides insight into the physiological roles of these molecules in animals.
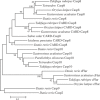
Figures




References
-
- Abraham MC, Shaham S. Death without caspases, caspases without death. Trends in Cell Biology. 2004;14:184–193. - PubMed
-
- Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nature Reviews Immunology. 2003;3:745–756. - PubMed
-
- Ahmad M, Srinivasula SM, Hegde R, Mukattash R, Fernandes-Alnemri T, Alnemri ES. Identification and characterization of murine caspase-14, a new member of the caspase family. Cancer Research. 1998;58:5201–5205. - PubMed
-
- Ashkenazi A, Dixit VM. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
