Cross-species comparison of aCGH data from mouse and human BRCA1- and BRCA2-mutated breast cancers
- PMID: 20735817
- PMCID: PMC2940799
- DOI: 10.1186/1471-2407-10-455
Cross-species comparison of aCGH data from mouse and human BRCA1- and BRCA2-mutated breast cancers
Abstract
Background: Genomic gains and losses are a result of genomic instability in many types of cancers. BRCA1- and BRCA2-mutated breast cancers are associated with increased amounts of chromosomal aberrations, presumably due their functions in genome repair. Some of these genomic aberrations may harbor genes whose absence or overexpression may give rise to cellular growth advantage. So far, it has not been easy to identify the driver genes underlying gains and losses. A powerful approach to identify these driver genes could be a cross-species comparison of array comparative genomic hybridization (aCGH) data from cognate mouse and human tumors. Orthologous regions of mouse and human tumors that are commonly gained or lost might represent essential genomic regions selected for gain or loss during tumor development.
Methods: To identify genomic regions that are associated with BRCA1- and BRCA2-mutated breast cancers we compared aCGH data from 130 mouse Brca1Δ/Δ;p53Δ/Δ, Brca2Δ/Δ;p53Δ/Δ and p53Δ/Δ mammary tumor groups with 103 human BRCA1-mutated, BRCA2-mutated and non-hereditary breast cancers.
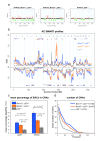
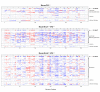
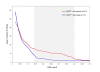
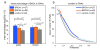
Results: Our genome-wide cross-species analysis yielded a complete collection of loci and genes that are commonly gained or lost in mouse and human breast cancer. Principal common CNAs were the well known MYC-associated gain and RB1/INTS6-associated loss that occurred in all mouse and human tumor groups, and the AURKA-associated gain occurred in BRCA2-related tumors from both species. However, there were also important differences between tumor profiles of both species, such as the prominent gain on chromosome 10 in mouse Brca2Δ/Δ;p53Δ/Δ tumors and the PIK3CA associated 3q gain in human BRCA1-mutated tumors, which occurred in tumors from one species but not in tumors from the other species. This disparity in recurrent aberrations in mouse and human tumors might be due to differences in tumor cell type or genomic organization between both species.
Conclusions: The selection of the oncogenome during mouse and human breast tumor development is markedly different, apart from the MYC gain and RB1-associated loss. These differences should be kept in mind when using mouse models for preclinical studies.
Figures



Similar articles
-
Prediction of BRCA2-association in hereditary breast carcinomas using array-CGH.Breast Cancer Res Treat. 2012 Apr;132(2):379-89. doi: 10.1007/s10549-010-1016-7. Epub 2010 Jul 8. Breast Cancer Res Treat. 2012. PMID: 20614180
-
Genomic subtypes of breast cancer identified by array-comparative genomic hybridization display distinct molecular and clinical characteristics.Breast Cancer Res. 2010;12(3):R42. doi: 10.1186/bcr2596. Epub 2010 Jun 24. Breast Cancer Res. 2010. PMID: 20576095 Free PMC article.
-
BRCA1-mutated and basal-like breast cancers have similar aCGH profiles and a high incidence of protein truncating TP53 mutations.BMC Cancer. 2010 Nov 30;10:654. doi: 10.1186/1471-2407-10-654. BMC Cancer. 2010. PMID: 21118481 Free PMC article.
-
Hereditary breast cancer; Genetic penetrance and current status with BRCA.J Cell Physiol. 2019 May;234(5):5741-5750. doi: 10.1002/jcp.27464. Epub 2018 Dec 14. J Cell Physiol. 2019. PMID: 30552672 Review.
-
The Tumor Suppressor BRCA1/2, Cancer Susceptibility and Genome Instability in Gynecological and Mammary Cancers.Asian Pac J Cancer Prev. 2023 Sep 1;24(9):3139-3153. doi: 10.31557/APJCP.2023.24.9.3139. Asian Pac J Cancer Prev. 2023. PMID: 37774066 Free PMC article. Review.
Cited by
-
Comparative oncogenomics identifies combinations of driver genes and drug targets in BRCA1-mutated breast cancer.Nat Commun. 2019 Jan 23;10(1):397. doi: 10.1038/s41467-019-08301-2. Nat Commun. 2019. PMID: 30674894 Free PMC article.
-
Mouse models in the era of large human tumour sequencing studies.Open Biol. 2018 Aug;8(8):180080. doi: 10.1098/rsob.180080. Open Biol. 2018. PMID: 30111589 Free PMC article. Review.
-
Genomic instability in breast and ovarian cancers: translation into clinical predictive biomarkers.Cell Mol Life Sci. 2012 Jan;69(2):223-45. doi: 10.1007/s00018-011-0809-0. Epub 2011 Sep 16. Cell Mol Life Sci. 2012. PMID: 21922196 Free PMC article. Review.
-
Lack of genomic heterogeneity at high-resolution aCGH between primary breast cancers and their paired lymph node metastases.PLoS One. 2014 Aug 1;9(8):e103177. doi: 10.1371/journal.pone.0103177. eCollection 2014. PLoS One. 2014. PMID: 25083859 Free PMC article.
-
BRCA1 promoter hypermethylation on circulating tumor DNA correlates with improved survival of patients with ovarian cancer.Mol Oncol. 2021 Dec;15(12):3615-3625. doi: 10.1002/1878-0261.13108. Epub 2021 Oct 12. Mol Oncol. 2021. PMID: 34601813 Free PMC article.
References
-
- Paakkonen K, Sauramo S, Sarantaus L, Vahteristo P, Hartikainen A, Vehmanen P, Ignatius J, Ollikainen V, Kaariainen H, Vauramo E. et al.Involvement of BRCA1 and BRCA2 in breast cancer in a western Finnish sub-population. Genet Epidemiol. 2001;20(2):239–246. doi: 10.1002/1098-2272(200102)20:2<239::AID-GEPI6>3.0.CO;2-Y. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

