Engineering of N. benthamiana L. plants for production of N-acetylgalactosamine-glycosylated proteins--towards development of a plant-based platform for production of protein therapeutics with mucin type O-glycosylation
- PMID: 20735851
- PMCID: PMC2936419
- DOI: 10.1186/1472-6750-10-62
Engineering of N. benthamiana L. plants for production of N-acetylgalactosamine-glycosylated proteins--towards development of a plant-based platform for production of protein therapeutics with mucin type O-glycosylation
Abstract
Background: Mucin type O-glycosylation is one of the most common types of post-translational modifications that impacts stability and biological functions of many mammalian proteins. A large family of UDP-GalNAc polypeptide:N-acetyl-α-galactosaminyltransferases (GalNAc-Ts) catalyzes the first step of mucin type O-glycosylation by transferring GalNAc to serine and/or threonine residues of acceptor polypeptides. Plants do not have the enzyme machinery to perform this process, thus restricting their use as bioreactors for production of recombinant therapeutic proteins.
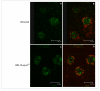
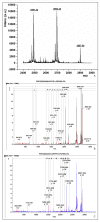
Results: The present study demonstrates that an isoform of the human GalNAc-Ts family, GalNAc-T2, retains its localization and functionality upon expression in N. benthamiana L. plants. The recombinant enzyme resides in the Golgi as evidenced by the fluorescence distribution pattern of the GalNAc-T2:GFP fusion and alteration of the fluorescence signature upon treatment with Brefeldin A. A GalNAc-T2-specific acceptor peptide, the 113-136 aa fragment of chorionic gonadotropin β-subunit, is glycosylated in vitro by the plant-produced enzyme at the "native" GalNAc attachment sites, Ser-121 and Ser-127. Ectopic expression of GalNAc-T2 is sufficient to "arm" tobacco cells with the ability to perform GalNAc-glycosylation, as evidenced by the attachment of GalNAc to Thr-119 of the endogenous enzyme endochitinase. However, glycosylation of highly expressed recombinant glycoproteins, like magnICON-expressed E. coli enterotoxin B subunit:H. sapiens mucin 1 tandem repeat-derived peptide fusion protein (LTBMUC1), is limited by the low endogenous UDP-GalNAc substrate pool and the insufficient translocation of UDP-GalNAc to the Golgi lumen. Further genetic engineering of the GalNAc-T2 plants by co-expressing Y. enterocolitica UDP-GlcNAc 4-epimerase gene and C. elegans UDP-GlcNAc/UDP-GalNAc transporter gene overcomes these limitations as indicated by the expression of the model LTBMUC1 protein exclusively as a glycoform.
Conclusion: Plant bioreactors can be engineered that are capable of producing Tn antigen-containing recombinant therapeutics.
Figures




Similar articles
-
Toward stable genetic engineering of human O-glycosylation in plants.Plant Physiol. 2012 Sep;160(1):450-63. doi: 10.1104/pp.112.198200. Epub 2012 Jul 12. Plant Physiol. 2012. PMID: 22791304 Free PMC article.
-
Engineering mammalian mucin-type O-glycosylation in plants.J Biol Chem. 2012 Apr 6;287(15):11911-23. doi: 10.1074/jbc.M111.312918. Epub 2012 Feb 14. J Biol Chem. 2012. PMID: 22334671 Free PMC article.
-
High level in vivo mucin-type glycosylation in Escherichia coli.Microb Cell Fact. 2018 Oct 26;17(1):168. doi: 10.1186/s12934-018-1013-9. Microb Cell Fact. 2018. PMID: 30367634 Free PMC article.
-
Control of mucin-type O-glycosylation: a classification of the polypeptide GalNAc-transferase gene family.Glycobiology. 2012 Jun;22(6):736-56. doi: 10.1093/glycob/cwr182. Epub 2011 Dec 18. Glycobiology. 2012. PMID: 22183981 Free PMC article. Review.
-
Myriad mechanisms: factors regulating the synthesis of aberrant mucin-type O-glycosylation found on cancer cells.Glycobiology. 2025 Apr 23;35(6):cwaf023. doi: 10.1093/glycob/cwaf023. Glycobiology. 2025. PMID: 40247681 Review.
Cited by
-
Challenges in O-glycan engineering of plants.Front Plant Sci. 2012 Sep 20;3:218. doi: 10.3389/fpls.2012.00218. eCollection 2012. Front Plant Sci. 2012. PMID: 23015807 Free PMC article.
-
Toward stable genetic engineering of human O-glycosylation in plants.Plant Physiol. 2012 Sep;160(1):450-63. doi: 10.1104/pp.112.198200. Epub 2012 Jul 12. Plant Physiol. 2012. PMID: 22791304 Free PMC article.
-
Transient co-expression with three O-glycosylation enzymes allows production of GalNAc-O-glycosylated Granulocyte-Colony Stimulating Factor in N. benthamiana.Plant Methods. 2018 Nov 6;14:98. doi: 10.1186/s13007-018-0363-y. eCollection 2018. Plant Methods. 2018. PMID: 30410568 Free PMC article.
-
N-glycosylation engineering of plants for the biosynthesis of glycoproteins with bisected and branched complex N-glycans.Glycobiology. 2011 Jun;21(6):813-23. doi: 10.1093/glycob/cwr009. Epub 2011 Feb 11. Glycobiology. 2011. PMID: 21317243 Free PMC article.
-
Advanced Plant-Based Glycan Engineering.Front Bioeng Biotechnol. 2018 Jun 14;6:81. doi: 10.3389/fbioe.2018.00081. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 29963553 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

