Ultrasensitive responses and specificity in cell signaling
- PMID: 20735856
- PMCID: PMC2940771
- DOI: 10.1186/1752-0509-4-119
Ultrasensitive responses and specificity in cell signaling
Abstract
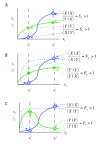
Background: Interconnected cell signaling pathways are able to efficiently and accurately transmit a multitude of different signals, despite an inherent potential for undesirable levels of cross-talk. To ensure that an appropriate response is produced, biological systems have evolved network-level mechanisms that insulate pathways from crosstalk and prevent 'leaking' or 'spillover' between pathways. Many signaling pathways have been shown to respond in an ultrasensitive (switch-like) fashion to graded input, and this behavior may influence specificity. The relationship of ultrasensitivity to signaling specificity has not been extensively explored.
Results: We studied the behavior of simple mathematical models of signaling networks composed of two interconnected pathways that share an intermediate component, asking if the two pathways in the network could exhibit both output specificity (preferentially activate their own output) and input fidelity (preferentially respond to their own input). Previous results with weakly-activated pathways indicated that neither mutual specificity nor mutual fidelity were obtainable in the absence of an insulating mechanism, such as cross-pathway inhibition, combinatorial signaling or scaffolding/compartmentalization. Here we found that mutual specificity is obtainable for hyperbolic or ultrasensitive pathways, even in the absence of an insulating mechanism. However, mutual fidelity is impossible at steady-state, even if pathways are hyperbolic or ultrasensitive. Nevertheless, ultrasensitivity does provide advantages in attaining specificity and fidelity to networks that contain an insulating mechanism. For networks featuring cross-pathway inhibition or combinatorial signaling, ultrasensitive activation can increase specificity in a limited way, and can only be utilized by one of the two pathways. In contrast, for networks featuring scaffolding/compartmentalization, ultrasensitive activation of both pathways can dramatically improve network specificity.
Conclusions: There are constraints to obtaining performance objectives associated with signaling specificity; such constraints may have influenced the evolution of signal transduction networks. Notably, input fidelity (preferential response to an authentic input) is a more difficult objective to achieve than output specificity (preferential targeting to an authentic output). Indeed, mutual fidelity is impossible in the absence of an insulating mechanism, even if pathways are ultrasensitive. Ultrasensitivity does, however, significantly enhance the performance of several insulating mechanisms. In particular, the ultrasensitive activation of both pathways can provide substantial improvement to networks containing scaffolding/compartmentalization.
Figures
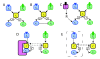
 . (B-E) Networks embellished with various insulating mechanisms (B) Cross Pathway Inhibition (CPI) from x2 to y2. (C) Combinatorial Signaling (CS) in the X pathway. (D) Scaffolding. (E) Compartmentalization.
. (B-E) Networks embellished with various insulating mechanisms (B) Cross Pathway Inhibition (CPI) from x2 to y2. (C) Combinatorial Signaling (CS) in the X pathway. (D) Scaffolding. (E) Compartmentalization.