A novel cytoplasmic adaptor for retinoic acid receptor (RAR) and thyroid receptor functions as a Derepressor of RAR in the absence of retinoic acid
- PMID: 20736163
- PMCID: PMC2962525
- DOI: 10.1074/jbc.M110.143008
A novel cytoplasmic adaptor for retinoic acid receptor (RAR) and thyroid receptor functions as a Derepressor of RAR in the absence of retinoic acid
Abstract
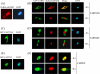
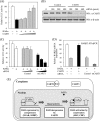
In most mammalian cells, the retinoic acid receptor (RAR) is nuclear rather than cytoplasmic, regardless of its cognate ligand, retinoic acid (RA). In testis Sertoli cells, however, RAR is retained in the cytoplasm and moves to the nucleus only when RA is supplied. This led us to identify a protein that regulates the translocation of RAR. From yeast two-hybrid screening, we identified a novel RAR-interacting protein called CART1 (cytoplasmic adaptor for RAR and TR). Systematic interaction assays using deletion mutants showed that the C-terminal CoRNR box of CART1 was responsible for the interaction with the NCoR binding region of RAR and TR. Such interaction was impaired in the presence of ligand RA, as further determined by GST pulldown assays in vitro and immunoprecipitation assays in vivo. Fluorescence microscopy showed that unliganded RAR was captured by CART1 in the cytoplasm, whereas liganded RAR was liberated and moved to the nucleus. Overexpression of CART1 blocked the transcriptional repressing activity of unliganded apoRAR, mediated by corepressor NCoR in the nucleus. CART1 siRNA treatment in a mouse Sertoli cell line, TM4, allowed RAR to move to the nucleus and blocked the derepressing function of CART1, suggesting that CART1 might be a cytoplasmic, testis-specific derepressor of RAR.
Figures





Similar articles
-
Regulation of the mouse preprothyrotropin-releasing hormone gene by retinoic acid receptor.Endocrinology. 1999 Nov;140(11):5004-13. doi: 10.1210/endo.140.11.7111. Endocrinology. 1999. PMID: 10537125
-
The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains.Mol Endocrinol. 2001 Jul;15(7):1049-61. doi: 10.1210/mend.15.7.0669. Mol Endocrinol. 2001. PMID: 11435607
-
Transcriptional silencing by unliganded thyroid hormone receptor beta requires a soluble corepressor that interacts with the ligand-binding domain of the receptor.Mol Cell Biol. 1996 May;16(5):1909-20. doi: 10.1128/MCB.16.5.1909. Mol Cell Biol. 1996. PMID: 8628257 Free PMC article.
-
Role of co-activators and co-repressors in the mechanism of steroid/thyroid receptor action.Recent Prog Horm Res. 1997;52:141-64; discussion 164-5. Recent Prog Horm Res. 1997. PMID: 9238851 Review.
-
Ligand-dependent interaction of nuclear receptors with potential transcriptional intermediary factors (mediators).Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):569-78. doi: 10.1098/rstb.1996.0056. Philos Trans R Soc Lond B Biol Sci. 1996. PMID: 8735280 Review.
Cited by
-
Is beta-carotene consumption associated with thyroid hormone levels?Front Endocrinol (Lausanne). 2023 May 26;14:1089315. doi: 10.3389/fendo.2023.1089315. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 37305054 Free PMC article. Review.
-
Nuclear receptors in health and disease: signaling pathways, biological functions and pharmaceutical interventions.Signal Transduct Target Ther. 2025 Jul 28;10(1):228. doi: 10.1038/s41392-025-02270-3. Signal Transduct Target Ther. 2025. PMID: 40717128 Free PMC article. Review.
-
Vitamin A, endocrine tissues and hormones: interplay and interactions.Endocr Connect. 2017 Aug 9;6(7):R121-R130. doi: 10.1530/EC-17-0101. Print 2017 Oct. Endocr Connect. 2017. PMID: 28720593 Free PMC article.
-
Diencephalic Size Is Restricted by a Novel Interplay Between GCN5 Acetyltransferase Activity and Retinoic Acid Signaling.J Neurosci. 2017 Mar 8;37(10):2565-2579. doi: 10.1523/JNEUROSCI.2121-16.2017. Epub 2017 Feb 2. J Neurosci. 2017. PMID: 28154153 Free PMC article.
-
Ubiquitin specific protease 19 involved in transcriptional repression of retinoic acid receptor by stabilizing CORO2A.Oncotarget. 2016 Jun 7;7(23):34759-72. doi: 10.18632/oncotarget.8976. Oncotarget. 2016. PMID: 27129179 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

