Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8·STAM complex
- PMID: 20736164
- PMCID: PMC2966105
- DOI: 10.1074/jbc.M109.016287
Regulation of epidermal growth factor receptor ubiquitination and trafficking by the USP8·STAM complex
Abstract
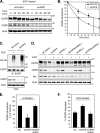
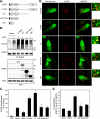
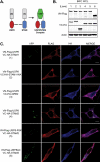
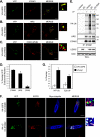
Reversible ubiquitination of activated receptor complexes signals their sorting between recycling and degradation and thereby dictates receptor fate. The deubiquitinating enzyme ubiquitin-specific protease 8 (USP8/UBPy) has been previously implicated in the regulation of the epidermal growth factor receptor (EGFR); however, the molecular mechanisms governing its recruitment and activity in this context remain unclear. Herein, we investigate the role of USP8 in countering ligand-induced ubiquitination and down-regulation of EGFR and characterize a subset of protein-protein interaction determinants critical for this function. USP8 depletion accelerates receptor turnover, whereas loss of hepatocyte growth factor-regulated substrate (Hrs) rescues this phenotype, indicating that USP8 protects EGFR from degradation via an Hrs-dependent pathway. Catalytic inactivation of USP8 incurs EGFR hyperubiquitination and promotes receptor localization to endosomes marked by high ubiquitin content. These phenotypes require the central region of USP8, containing three extended Arg-X-X-Lys (RXXK) motifs that specify direct low affinity interactions with the SH3 domain(s) of ESCRT-0 proteins, STAM1/2. The USP8·STAM complex critically impinges on receptor ubiquitination status and modulates ubiquitin dynamics on EGFR-positive endosomes. Consequently, USP8-mediated deubiquitination slows progression of EGFR past the early-to-recycling endosome circuit in a manner dependent upon the RXXK motifs. Collectively, these findings demonstrate a role for the USP8·STAM complex as a protective mechanism regulating early endosomal sorting of EGFR between pathways destined for lysosomal degradation and recycling.
Figures

References
-
- Polo S., Di Fiore P. P. (2006) Cell 124, 897–900 - PubMed
-
- Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 - PubMed
-
- Haglund K., Sigismund S., Polo S., Szymkiewicz I., Di Fiore P. P., Dikic I. (2003) Nat. Cell Biol. 5, 461–466 - PubMed
-
- Hicke L., Riezman H. (1996) Cell 84, 277–287 - PubMed
-
- Duan L., Miura Y., Dimri M., Majumder B., Dodge I. L., Reddi A. L., Ghosh A., Fernandes N., Zhou P., Mullane-Robinson K., Rao N., Donoghue S., Rogers R. A., Bowtell D., Naramura M., Gu H., Band V., Band H. (2003) J. Biol. Chem. 278, 28950–28960 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

