Direct association of Sprouty-related protein with an EVH1 domain (SPRED) 1 or SPRED2 with DYRK1A modifies substrate/kinase interactions
- PMID: 20736167
- PMCID: PMC2975161
- DOI: 10.1074/jbc.M110.148445
Direct association of Sprouty-related protein with an EVH1 domain (SPRED) 1 or SPRED2 with DYRK1A modifies substrate/kinase interactions
Abstract
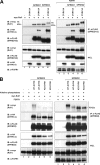
The mammalian SPRED (Sprouty-related protein with an EVH1 domain) proteins include a family of three members, SPRED1-3. Currently, little is known about their biochemistry. The best described, SPRED1, has been shown to inhibit the Ras/ERK pathway downstream of Ras. All three SPREDs have a cysteine-rich domain (CRD) that has high homology to the CRD of the Sprouty family of proteins, several of which are also Ras/ERK inhibitors. In the belief that binding partners would clarify SPRED function, we assayed for their associated proteins. Here, we describe the direct and endogenous interaction of SPRED1 and SPRED2 with the novel kinase, DYRK1A. DYRK1A has become the subject of recent research focus as it plays a central role in Caenorhabditis elegans oocyte maturation and egg activation, and there is strong evidence that it could be involved in Down syndrome in humans. Both SPRED1 and SPRED2 inhibit the ability of DYRK1A to phosphorylate its substrates, Tau and STAT3. This inhibition occurs via an interaction of the CRD of the SPREDs with the kinase domain of DYRK1A. DYRK1A substrates must bind to the kinase to enable phosphorylation, and SPRED proteins compete for the same binding site to modify this process. Our accumulated evidence indicates that the SPRED proteins are likely physiological modifiers of DYRK1A.
Figures



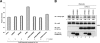
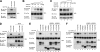


Similar articles
-
SPREDs (Sprouty related proteins with EVH1 domain) promote self-renewal and inhibit mesodermal differentiation in murine embryonic stem cells.Dev Dyn. 2015 Apr;244(4):591-606. doi: 10.1002/dvdy.24261. Epub 2015 Mar 13. Dev Dyn. 2015. PMID: 25690936
-
Sprouty-related Ena/vasodilator-stimulated phosphoprotein homology 1-domain-containing protein (SPRED1), a tyrosine-protein phosphatase non-receptor type 11 (SHP2) substrate in the Ras/extracellular signal-regulated kinase (ERK) pathway.J Biol Chem. 2011 Jul 1;286(26):23102-12. doi: 10.1074/jbc.M110.212662. Epub 2011 Apr 29. J Biol Chem. 2011. PMID: 21531714 Free PMC article.
-
The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization.Oncogene. 2004 Jul 22;23(33):5567-76. doi: 10.1038/sj.onc.1207759. Oncogene. 2004. PMID: 15184877
-
Sprouty proteins: modified modulators, matchmakers or missing links?J Endocrinol. 2009 Nov;203(2):191-202. doi: 10.1677/JOE-09-0110. Epub 2009 May 7. J Endocrinol. 2009. PMID: 19423641 Review.
-
Progress in experimental research on SPRED protein family.J Int Med Res. 2020 Aug;48(8):300060520929170. doi: 10.1177/0300060520929170. J Int Med Res. 2020. PMID: 32851895 Free PMC article. Review.
Cited by
-
Non-synonymous, synonymous, and non-coding nucleotide variants contribute to recurrently altered biological processes during retinoblastoma progression.Genes Chromosomes Cancer. 2023 May;62(5):275-289. doi: 10.1002/gcc.23120. Epub 2023 Jan 5. Genes Chromosomes Cancer. 2023. PMID: 36550020 Free PMC article.
-
DYRK1A overexpression enhances STAT activity and astrogliogenesis in a Down syndrome mouse model.EMBO Rep. 2015 Nov;16(11):1548-62. doi: 10.15252/embr.201540374. Epub 2015 Sep 15. EMBO Rep. 2015. PMID: 26373433 Free PMC article.
-
Identification of FAM53C as a cytosolic-anchoring inhibitory binding protein of the kinase DYRK1A.Life Sci Alliance. 2023 Oct 6;6(12):e202302129. doi: 10.26508/lsa.202302129. Print 2023 Dec. Life Sci Alliance. 2023. PMID: 37802655 Free PMC article.
-
Receptor tyrosine kinase (RTK) signalling in the control of neural stem and progenitor cell (NSPC) development.Mol Neurobiol. 2014 Feb;49(1):440-71. doi: 10.1007/s12035-013-8532-5. Epub 2013 Aug 28. Mol Neurobiol. 2014. PMID: 23982746 Review.
-
A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1.Genes Dev. 2012 Jul 1;26(13):1421-6. doi: 10.1101/gad.190876.112. Genes Dev. 2012. PMID: 22751498 Free PMC article.
References
-
- Schlessinger J. (2000) Cell 103, 211–225 - PubMed
-
- Schlessinger J. (2002) Cell 110, 669–672 - PubMed
-
- Zwick E., Bange J., Ullrich A. (2001) Endocr.-Relat. Cancer 8, 161–173 - PubMed
-
- Shaw R. J., Cantley L. C. (2006) Nature 441, 424–430 - PubMed
-
- Midgley R. S., Kerr D. J. (2002) Crit. Rev. Oncol. Hematol. 44, 109–120 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

