Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB)
- PMID: 20736178
- PMCID: PMC2966045
- DOI: 10.1074/jbc.M110.162925
Double strand break unwinding and resection by the mycobacterial helicase-nuclease AdnAB in the presence of single strand DNA-binding protein (SSB)
Abstract
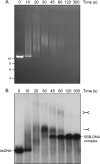
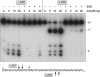
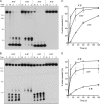
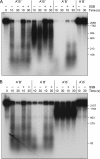
Mycobacterial AdnAB is a heterodimeric DNA helicase-nuclease and 3' to 5' DNA translocase implicated in the repair of double strand breaks (DSBs). The AdnA and AdnB subunits are each composed of an N-terminal motor domain and a C-terminal nuclease domain. Inclusion of mycobacterial single strand DNA-binding protein (SSB) in reactions containing linear plasmid dsDNA allowed us to study the AdnAB helicase under conditions in which the unwound single strands are coated by SSB and thereby prevented from reannealing or promoting ongoing ATP hydrolysis. We found that the AdnAB motor catalyzed processive unwinding of 2.7-11.2-kbp linear duplex DNAs at a rate of ∼250 bp s(-1), while hydrolyzing ∼5 ATPs per bp unwound. Crippling the AdnA phosphohydrolase active site did not affect the rate of unwinding but lowered energy consumption slightly, to ∼4.2 ATPs bp(-1). Mutation of the AdnB phosphohydrolase abolished duplex unwinding, consistent with a model in which the "leading" AdnB motor propagates a Y-fork by translocation along the 3' DNA strand, ahead of the "lagging" AdnA motor domain. By tracking the resection of the 5' and 3' strands at the DSB ends, we illuminated a division of labor among the AdnA and AdnB nuclease modules during dsDNA unwinding, whereby the AdnA nuclease processes the unwound 5' strand to liberate a short oligonucleotide product, and the AdnB nuclease incises the 3' strand on which the motor translocates. These results extend our understanding of presynaptic DSB processing by AdnAB and engender instructive comparisons with the RecBCD and AddAB clades of bacterial helicase-nuclease machines.
Figures
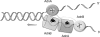





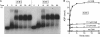

Similar articles
-
Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24507-24516. doi: 10.1073/pnas.1913546116. Epub 2019 Nov 18. Proc Natl Acad Sci U S A. 2019. PMID: 31740608 Free PMC article.
-
Characterization of the mycobacterial AdnAB DNA motor provides insights into the evolution of bacterial motor-nuclease machines.J Biol Chem. 2010 Jan 22;285(4):2632-41. doi: 10.1074/jbc.M109.076133. Epub 2009 Nov 17. J Biol Chem. 2010. PMID: 19920138 Free PMC article.
-
AdnAB: a new DSB-resecting motor-nuclease from mycobacteria.Genes Dev. 2009 Jun 15;23(12):1423-37. doi: 10.1101/gad.1805709. Epub 2009 May 26. Genes Dev. 2009. PMID: 19470566 Free PMC article.
-
The processing of double-stranded DNA breaks for recombinational repair by helicase-nuclease complexes.DNA Repair (Amst). 2010 Mar 2;9(3):276-85. doi: 10.1016/j.dnarep.2009.12.016. Epub 2010 Jan 29. DNA Repair (Amst). 2010. PMID: 20116346 Review.
-
Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB.Nat Rev Microbiol. 2013 Jan;11(1):9-13. doi: 10.1038/nrmicro2917. Epub 2012 Dec 3. Nat Rev Microbiol. 2013. PMID: 23202527 Review.
Cited by
-
Mycobacterium smegmatis RqlH defines a novel clade of bacterial RecQ-like DNA helicases with ATP-dependent 3'-5' translocase and duplex unwinding activities.Nucleic Acids Res. 2012 May;40(10):4604-14. doi: 10.1093/nar/gks046. Epub 2012 Jan 28. Nucleic Acids Res. 2012. PMID: 22287622 Free PMC article.
-
RecF and RecR Play Critical Roles in the Homologous Recombination and Single-Strand Annealing Pathways of Mycobacteria.J Bacteriol. 2015 Oct;197(19):3121-32. doi: 10.1128/JB.00290-15. Epub 2015 Jul 20. J Bacteriol. 2015. PMID: 26195593 Free PMC article.
-
Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.Proc Natl Acad Sci U S A. 2019 Dec 3;116(49):24507-24516. doi: 10.1073/pnas.1913546116. Epub 2019 Nov 18. Proc Natl Acad Sci U S A. 2019. PMID: 31740608 Free PMC article.
-
Homologous recombination mediated by the mycobacterial AdnAB helicase without end resection by the AdnAB nucleases.Nucleic Acids Res. 2017 Jan 25;45(2):762-774. doi: 10.1093/nar/gkw1130. Epub 2016 Nov 29. Nucleic Acids Res. 2017. PMID: 27899634 Free PMC article.
-
Mycobacterium smegmatis HelY Is an RNA-Activated ATPase/dATPase and 3'-to-5' Helicase That Unwinds 3'-Tailed RNA Duplexes and RNA:DNA Hybrids.J Bacteriol. 2015 Oct;197(19):3057-65. doi: 10.1128/JB.00418-15. Epub 2015 Jul 13. J Bacteriol. 2015. PMID: 26170411 Free PMC article.
References
-
- Singleton M. R., Dillingham M. S., Gaudier M., Kowalczykowski S. C., Wigley D. B. (2004) Nature 432, 187–193 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

