Reshaping the gut microbiome with bacterial transplantation and antibiotic intake
- PMID: 20736229
- PMCID: PMC2945190
- DOI: 10.1101/gr.107987.110
Reshaping the gut microbiome with bacterial transplantation and antibiotic intake
Abstract
The intestinal microbiota consists of over 1000 species, which play key roles in gut physiology and homeostasis. Imbalances in the composition of this bacterial community can lead to transient intestinal dysfunctions and chronic disease states. Understanding how to manipulate this ecosystem is thus essential for treating many disorders. In this study, we took advantage of recently developed tools for deep sequencing and phylogenetic clustering to examine the long-term effects of exogenous microbiota transplantation combined with and without an antibiotic pretreatment. In our rat model, deep sequencing revealed an intestinal bacterial diversity exceeding that of the human gut by a factor of two to three. The transplantation produced a marked increase in the microbial diversity of the recipients, which stemmed from both capture of new phylotypes and increase in abundance of others. However, when transplantation was performed after antibiotic intake, the resulting state simply combined the reshaping effects of the individual treatments (including the reduced diversity from antibiotic treatment alone). Therefore, lowering the recipient bacterial load by antibiotic intake prior to transplantation did not increase establishment of the donor phylotypes, although some dominant lineages still transferred successfully. Remarkably, all of these effects were observed after 1 mo of treatment and persisted after 3 mo. Overall, our results indicate that the indigenous gut microbial composition is more plastic that previously anticipated. However, since antibiotic pretreatment counterintuitively interferes with the establishment of an exogenous community, such plasticity is likely conditioned more by the altered microbiome gut homeostasis caused by antibiotics than by the primary bacterial loss.
Figures
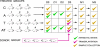
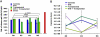


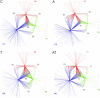
References
-
- Alpert C, Sczesny S, Gruhl B, Blaut M 2008. Long-term stability of the human gut microbiota in two different rat strains. Curr Issues Mol Biol 10: 17–24 - PubMed
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI 2005. Host-bacterial mutualism in the human intestine. Science 307: 1915–1920 - PubMed
-
- Borody TJ, Warren EF, Leis S, Surace R, Ashman O 2003. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol 37: 42–47 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
