HLH54F is required for the specification and migration of longitudinal gut muscle founders from the caudal mesoderm of Drosophila
- PMID: 20736287
- PMCID: PMC2926959
- DOI: 10.1242/dev.046573
HLH54F is required for the specification and migration of longitudinal gut muscle founders from the caudal mesoderm of Drosophila
Abstract
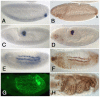
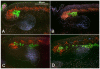
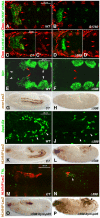
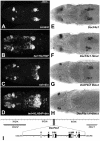
HLH54F, the Drosophila ortholog of the vertebrate basic helix-loop-helix domain-encoding genes capsulin and musculin, is expressed in the founder cells and developing muscle fibers of the longitudinal midgut muscles. These cells descend from the posterior-most portion of the mesoderm, termed the caudal visceral mesoderm (CVM), and migrate onto the trunk visceral mesoderm prior to undergoing myoblast fusion and muscle fiber formation. We show that HLH54F expression in the CVM is regulated by a combination of terminal patterning genes and snail. We generated HLH54F mutations and show that this gene is crucial for the specification, migration and survival of the CVM cells and the longitudinal midgut muscle founders. HLH54F mutant embryos, larvae, and adults lack all longitudinal midgut muscles, which causes defects in gut morphology and integrity. The function of HLH54F as a direct activator of gene expression is exemplified by our analysis of a CVM-specific enhancer from the Dorsocross locus, which requires combined inputs from HLH54F and Biniou in a feed-forward fashion. We conclude that HLH54F is the earliest specific regulator of CVM development and that it plays a pivotal role in all major aspects of development and differentiation of this largely twist-independent population of mesodermal cells.
Figures




Similar articles
-
The role of FGF signaling in guiding coordinate movement of cell groups: guidance cue and cell adhesion regulator?Cell Adh Migr. 2012 Sep-Oct;6(5):397-403. doi: 10.4161/cam.21103. Epub 2012 Sep 1. Cell Adh Migr. 2012. PMID: 23076054 Free PMC article. Review.
-
Hand is a direct target of the forkhead transcription factor Biniou during Drosophila visceral mesoderm differentiation.BMC Dev Biol. 2007 May 18;7:49. doi: 10.1186/1471-213X-7-49. BMC Dev Biol. 2007. PMID: 17511863 Free PMC article.
-
The FGF8-related signals Pyramus and Thisbe promote pathfinding, substrate adhesion, and survival of migrating longitudinal gut muscle founder cells.Dev Biol. 2012 Aug 1;368(1):28-43. doi: 10.1016/j.ydbio.2012.05.010. Epub 2012 May 17. Dev Biol. 2012. PMID: 22609944 Free PMC article.
-
Synchronous and symmetric migration of Drosophila caudal visceral mesoderm cells requires dual input by two FGF ligands.Development. 2012 Feb;139(4):699-708. doi: 10.1242/dev.068791. Epub 2012 Jan 4. Development. 2012. PMID: 22219352 Free PMC article.
-
Determination and development of the larval muscle pattern in Drosophila melanogaster.Cell Tissue Res. 1999 Apr;296(1):151-60. doi: 10.1007/s004410051276. Cell Tissue Res. 1999. PMID: 10199975 Review.
Cited by
-
Single-cell transcriptomics illuminates regulatory steps driving anterior-posterior patterning of Drosophila embryonic mesoderm.Cell Rep. 2023 Oct 31;42(10):113289. doi: 10.1016/j.celrep.2023.113289. Epub 2023 Oct 19. Cell Rep. 2023. PMID: 37858470 Free PMC article.
-
The role of FGF signaling in guiding coordinate movement of cell groups: guidance cue and cell adhesion regulator?Cell Adh Migr. 2012 Sep-Oct;6(5):397-403. doi: 10.4161/cam.21103. Epub 2012 Sep 1. Cell Adh Migr. 2012. PMID: 23076054 Free PMC article. Review.
-
Origin, Specification, and Plasticity of the Great Vessels of the Heart.Curr Biol. 2015 Aug 17;25(16):2099-110. doi: 10.1016/j.cub.2015.06.076. Epub 2015 Aug 6. Curr Biol. 2015. PMID: 26255850 Free PMC article.
-
Two sequential gene expression programs bridged by cell division support long-distance collective cell migration.Development. 2024 May 15;151(10):dev202262. doi: 10.1242/dev.202262. Epub 2024 May 17. Development. 2024. PMID: 38646822 Free PMC article.
-
A large reverse-genetic screen identifies numerous regulators of testis nascent myotube collective cell migration and collective organ sculpting.Mol Biol Cell. 2025 Feb 1;36(2):ar21. doi: 10.1091/mbc.E24-10-0456. Epub 2025 Jan 2. Mol Biol Cell. 2025. PMID: 39745864 Free PMC article.
References
-
- Azpiazu N., Frasch M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340 - PubMed
-
- Barolo S., Carver L. A., Posakony J. W. (2000). GFP and β-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29, 726-732 - PubMed
-
- Bonini N. M., Fortini M. E. (1999). Surviving Drosophila eye development: integrating cell death with differentiation during formation of a neural structure. BioEssays 21, 991-1003 - PubMed
-
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

