Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants
- PMID: 20736351
- PMCID: PMC2941279
- DOI: 10.1073/pnas.1007926107
Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants
Abstract
The growth of an organism and its size determination require the tight regulation of cell proliferation and cell growth. However, the mechanisms and regulatory networks that control and integrate these processes remain poorly understood. Here, we address the biological role of Arabidopsis translationally controlled tumor protein (AtTCTP) and test its shared functions in animals and plants. The data support a role of plant AtTCTP as a positive regulator of mitotic growth by specifically controlling the duration of the cell cycle. We show that, in contrast to animal TCTP, plant AtTCTP is not implicated in regulating postmitotic growth. Consistent with this finding, plant AtTCTP can fully rescue cell proliferation defects in Drosophila loss of function for dTCTP. Furthermore, Drosophila dTCTP is able to fully rescue cell proliferation defects in Arabidopsis tctp knockouts. Our data provide evidence that TCTP function in regulating cell division is part of a conserved growth regulatory pathway shared between plants and animals. The study also suggests that, although the cell division machinery is shared in all multicellular organisms to control growth, cell expansion can be uncoupled from cell division in plants but not in animals.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
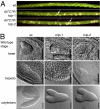
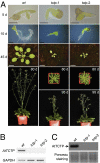
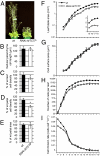
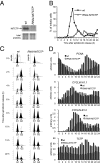
References
-
- Busov VB, Brunner AM, Strauss SH. Genes for control of plant stature and form. New Phytol. 2008;177:589–607. - PubMed
-
- Krizek BA. Making bigger plants: Key regulators of final organ size. Curr Opin Plant Biol. 2009;12:17–22. - PubMed
-
- Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. - PubMed
-
- Bommer UA, Thiele BJ. The translationally controlled tumour protein (TCTP) Int J Biochem Cell Biol. 2004;36:379–385. - PubMed
-
- Hinojosa-Moya J, et al. Phylogenetic and structural analysis of translationally controlled tumor proteins. J Mol Evol. 2008;66:472–483. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

