Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus
- PMID: 20736368
- PMCID: PMC3031132
- DOI: 10.1158/0008-5472.CAN-10-0765
Aberrant silencing of cancer-related genes by CpG hypermethylation occurs independently of their spatial organization in the nucleus
Abstract
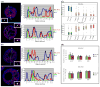
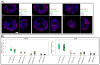
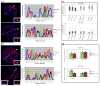
Aberrant promoter DNA-hypermethylation and repressive chromatin constitutes a frequent mechanism of gene inactivation in cancer. There is great interest in dissecting the mechanisms underlying this abnormal silencing. Studies have shown changes in the nuclear organization of chromatin in tumor cells as well as the association of aberrant methylation with long-range silencing of neighboring genes. Furthermore, certain tumors show a high incidence of promoter methylation termed as the CpG island methylator phenotype. Here, we have analyzed the role of nuclear chromatin architecture for genes in hypermethylated inactive versus nonmethylated active states and its relation with long-range silencing and CpG island methylator phenotype. Using combined immunostaining for active/repressive chromatin marks and fluorescence in situ hybridization in colorectal cancer cell lines, we show that aberrant silencing of these genes occurs without requirement for their being positioned at heterochromatic domains. Importantly, hypermethylation, even when associated with long-range epigenetic silencing of neighboring genes, occurs independent of their euchromatic or heterochromatic location. Together, these results indicate that, in cancer, extensive changes around promoter chromatin of individual genes or gene clusters could potentially occur locally without preference for nuclear position and/or causing repositioning. These findings have important implications for understanding relationships between nuclear organization and gene expression patterns in cancer.
Figures


Similar articles
-
Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma.Clin Cancer Res. 2008 May 1;14(9):2560-9. doi: 10.1158/1078-0432.CCR-07-1802. Clin Cancer Res. 2008. PMID: 18451217 Free PMC article.
-
MGMT and MLH1 promoter methylation versus APC, KRAS and BRAF gene mutations in colorectal cancer: indications for distinct pathways and sequence of events.Ann Oncol. 2009 Jul;20(7):1216-22. doi: 10.1093/annonc/mdn782. Epub 2009 Jan 22. Ann Oncol. 2009. PMID: 19164452
-
Promoter hypermethylation of DNA damage response genes in hepatocellular carcinoma.Cell Biol Int. 2012 May 1;36(5):427-32. doi: 10.1042/CBI20100851. Cell Biol Int. 2012. PMID: 21864295
-
CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future.Oncogene. 2002 Aug 12;21(35):5427-40. doi: 10.1038/sj.onc.1205600. Oncogene. 2002. PMID: 12154405 Review.
-
Epigenetic gene silencing in cancer initiation and progression.Cancer Lett. 2003 Feb 20;190(2):125-33. doi: 10.1016/s0304-3835(02)00511-6. Cancer Lett. 2003. PMID: 12565166 Review.
Cited by
-
FastDMA: an infinium humanmethylation450 beadchip analyzer.PLoS One. 2013 Sep 5;8(9):e74275. doi: 10.1371/journal.pone.0074275. eCollection 2013. PLoS One. 2013. PMID: 24040221 Free PMC article.
-
Role of nuclear architecture in epigenetic alterations in cancer.Cold Spring Harb Symp Quant Biol. 2010;75:507-15. doi: 10.1101/sqb.2010.75.031. Epub 2011 Mar 29. Cold Spring Harb Symp Quant Biol. 2010. PMID: 21447817 Free PMC article.
-
Neuropeptide receptor genes GHSR and NMUR1 are candidate epigenetic biomarkers and predictors for surgically treated patients with oropharyngeal cancer.Sci Rep. 2020 Jan 23;10(1):1007. doi: 10.1038/s41598-020-57920-z. Sci Rep. 2020. PMID: 31974445 Free PMC article.
-
Cancer epigenetics: linking basic biology to clinical medicine.Cell Res. 2011 Mar;21(3):502-17. doi: 10.1038/cr.2011.24. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321605 Free PMC article. Review.
-
Genome-wide positioning of bivalent mononucleosomes.BMC Med Genomics. 2016 Sep 15;9(1):60. doi: 10.1186/s12920-016-0221-6. BMC Med Genomics. 2016. PMID: 27634286 Free PMC article.
References
-
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–9. - PubMed
-
- Bender CM, Pao MM, Jones PA. Inhibition of DNA methylation by 5-aza-2′-deoxycytidine suppresses the growth of human tumor cell lines. Cancer Res. 1998;58:95–101. - PubMed
-
- Egger G, Aparicio AM, Escobar SG, Jones PA. Inhibition of histone deacetylation does not block resilencing of p16 after 5-aza-2′-deoxycytidine treatment. Cancer Res. 2007;67:346–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

