The impact of C-MYC gene expression on gastric cancer cell
- PMID: 20737197
- PMCID: PMC3618981
- DOI: 10.1007/s11010-010-0536-0
The impact of C-MYC gene expression on gastric cancer cell
Abstract
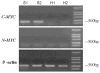
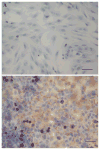
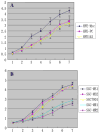
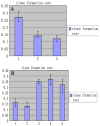
The upregulation or mutation of C-MYC has been observed in gastric, colon, breast, and lung tumors and in Burkitt's lymphoma. However, little is known about the role C-MYC plays in gastric adenocarcinoma. In the present study, we intended to investigate the influence of C-MYC on the growth, proliferation, apoptosis, invasion, and cell cycle of the gastric cancer cell line SGC7901 and the gastric cell line HFE145. C-MYC cDNA was subcloned into a constitutive vector PCDNA3.1 followed by transfection in normal gastric cell line HFE145 by using liposome. Then stable transfectants were selected and appraised. Specific inhibition of C-MYC was achieved using a vector-based siRNA system which was transfected in gastric cancer cell line SGC7901. The apoptosis and cell cycles of these clones were analyzed by using flow cytometric assay. The growth and proliferation were analyzed by cell growth curves and colony-forming assay, respectively. The invasion of these clones was analyzed by using cell migration assay. The C-MYC stable expression clones (HFE-Myc) and C-MYC RNAi cells (SGC-MR) were detected and compared with their control groups, respectively. HFE-Myc grew faster than HFE145 and HFE-PC (HFE145 transfected with PCDNA3.1 vector). SGC-MR1, 2 grew slower than SGC7901 and SGC-MS1, 2 (SGC7901 transfected with scrambled control duplexes). The cell counts of HFE-Myc in the third, fourth, fifth, sixth, and seventh days were significantly more than those of control groups (P < 0.05). Those of SGC-MR1, 2 in the fourth, fifth, sixth, and seventh days were significantly fewer than those of control groups (P < 0.05). Cell cycle analysis showed that proportions of HFE-Myc and SGC-MR cells in G0-G1 and G2-M were different significantly with their control groups, respectively (P < 0.05). The apoptosis rate of HFE-Myc was significantly higher than those of control groups (P < 0.05). Results of colony-forming assay showed that the colony formation rate of HFE-Myc was higher than those of control groups; otherwise, the rate of SGC-MR was lower than those of their control groups (P < 0.05). The results of cell migration assay showed that there were no significant differences between experimental groups and control groups (P > 0.05). In conclusion, C-MYC can promote the growth and proliferation of normal gastric cells, and knockdown of C-MYC can restrain the growth and proliferation of gastric cancer cells. It can induce cell apoptosis and help tumor cell maintain malignant phenotype. But it can have not a detectable influence on the ability of invasion of gastric cancer cells.
Conflict of interest statement
Figures





Similar articles
-
The influence of TXNDC5 gene on gastric cancer cell.J Cancer Res Clin Oncol. 2010 Oct;136(10):1497-505. doi: 10.1007/s00432-010-0807-x. Epub 2010 Feb 16. J Cancer Res Clin Oncol. 2010. PMID: 20157732 Free PMC article.
-
A study on the functions of ubiquitin metabolic system related gene FBG2 in gastric cancer cell line.J Exp Clin Cancer Res. 2009 Jun 10;28(1):78. doi: 10.1186/1756-9966-28-78. J Exp Clin Cancer Res. 2009. PMID: 19515249 Free PMC article.
-
Impact of BTG2 expression on proliferation and invasion of gastric cancer cells in vitro.Mol Biol Rep. 2010 Jul;37(6):2579-86. doi: 10.1007/s11033-009-9777-y. Epub 2009 Sep 2. Mol Biol Rep. 2010. PMID: 19728149
-
The role of calcyclin gene in the proliferation and migration of gastric cancer cells.Oncol Rep. 2011 Sep 20. doi: 10.3892/or.2011.1467. Online ahead of print. Oncol Rep. 2011. PMID: 21935579
-
Function of chloride intracellular channel 1 in gastric cancer cells.World J Gastroenterol. 2012 Jun 28;18(24):3070-80. doi: 10.3748/wjg.v18.i24.3070. World J Gastroenterol. 2012. PMID: 22791942 Free PMC article.
Cited by
-
Gastric Cancer Cell Lines Have Different MYC-Regulated Expression Patterns but Share a Common Core of Altered Genes.Can J Gastroenterol Hepatol. 2018 Oct 16;2018:5804376. doi: 10.1155/2018/5804376. eCollection 2018. Can J Gastroenterol Hepatol. 2018. PMID: 30410872 Free PMC article.
-
MicroRNA-27b inhibits the development of melanoma by targeting MYC.Oncol Lett. 2021 May;21(5):370. doi: 10.3892/ol.2021.12631. Epub 2021 Mar 12. Oncol Lett. 2021. PMID: 33747226 Free PMC article.
-
Copy number alterations and epithelial‑mesenchymal transition genes in diffuse and intestinal gastric cancers in Mexican patients.Mol Med Rep. 2022 May;25(5):191. doi: 10.3892/mmr.2022.12707. Epub 2022 Apr 1. Mol Med Rep. 2022. PMID: 35362543 Free PMC article.
-
Comprehensive multi-omics analysis of tryptophan metabolism-related gene expression signature to predict prognosis in gastric cancer.Front Pharmacol. 2023 Oct 16;14:1267186. doi: 10.3389/fphar.2023.1267186. eCollection 2023. Front Pharmacol. 2023. PMID: 37908977 Free PMC article.
-
The cancer/testis antigen HORMAD1 promotes gastric cancer progression by activating the NF-κB signaling pathway and inducing epithelial-mesenchymal transition.Am J Transl Res. 2023 Sep 15;15(9):5808-5825. eCollection 2023. Am J Transl Res. 2023. PMID: 37854207 Free PMC article.
References
-
- Parkin DM, Poisani P, Ferlay Global cancer statistics. CA Cancer J Clin. 1999;49(1):33–64. - PubMed
-
- Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—First American Cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52(24):6735–6740. - PubMed
-
- Gonzalez CA, Sala N, Capella G. Genetic susceptibility and gastric cancer risk. Int J Cancer. 2002;100(3):249–260. - PubMed
-
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

