The apicomplexan Cryptosporidium parvum possesses a single mitochondrial-type ferredoxin and ferredoxin:NADP+ reductase system
- PMID: 20737579
- PMCID: PMC3005779
- DOI: 10.1002/pro.487
The apicomplexan Cryptosporidium parvum possesses a single mitochondrial-type ferredoxin and ferredoxin:NADP+ reductase system
Abstract
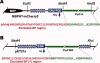
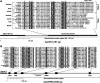
We have successfully expressed recombinant mitochondrial-type ferredoxin (mtFd) and ferredoxin:NADP(+) reductase (mtFNR) from Cryptosporidium parvum and characterized their biochemical features for the first time for an apicomplexan. Both C. parvum mtFd (CpmtFd) and FNR (CpmtFNR) were obtained and purified as holo-proteins, in which the correct assembly of [2Fe-2S] cluster in Fd and that of FAD in FNR were confirmed and characterized by UV/vis and electron paramagnetic resonance. These proteins were fully functional and CpmtFNR was capable of transferring electrons from NADPH to CpmtFd in a cytochrome c-coupled assay that followed a typical Michaelis-Menten kinetics. Apicomplexan mtFd and mtFNR proteins were evolutionarily divergent from their counterparts in humans and animals and could be explored as potential drug targets in Cryptosporidium and other apicomplexans.
Figures





Similar articles
-
The ferredoxin-NADP+ reductase/ferredoxin electron transfer system of Plasmodium falciparum.FEBS J. 2009 Jul;276(14):3825-36. doi: 10.1111/j.1742-4658.2009.07100.x. Epub 2009 Jun 11. FEBS J. 2009. PMID: 19523113
-
Identification and molecular characterization of mitochondrial ferredoxins and ferredoxin reductase from Arabidopsis.Plant Mol Biol. 2003 Jul;52(4):817-30. doi: 10.1023/a:1025015811141. Plant Mol Biol. 2003. PMID: 13677469
-
Ferredoxin-NADP+ reductase and ferredoxin of the protozoan parasite Toxoplasma gondii interact productively in Vitro and in Vivo.J Biol Chem. 2002 Dec 13;277(50):48463-71. doi: 10.1074/jbc.M209388200. Epub 2002 Oct 4. J Biol Chem. 2002. PMID: 12370173
-
Structural and functional diversity of ferredoxin-NADP(+) reductases.Arch Biochem Biophys. 2008 Jun 15;474(2):283-91. doi: 10.1016/j.abb.2008.02.014. Epub 2008 Feb 16. Arch Biochem Biophys. 2008. PMID: 18307973 Review.
-
Interaction and electron transfer between ferredoxin-NADP+ oxidoreductase and its partners: structural, functional, and physiological implications.Photosynth Res. 2017 Dec;134(3):265-280. doi: 10.1007/s11120-017-0372-0. Epub 2017 Mar 30. Photosynth Res. 2017. PMID: 28361449 Review.
Cited by
-
Validation of IMP dehydrogenase inhibitors in a mouse model of cryptosporidiosis.Antimicrob Agents Chemother. 2014;58(3):1603-14. doi: 10.1128/AAC.02075-13. Epub 2013 Dec 23. Antimicrob Agents Chemother. 2014. PMID: 24366728 Free PMC article.
-
Identification of Sequences Encoding Symbiodinium minutum Mitochondrial Proteins.Genome Biol Evol. 2016 Jan 21;8(2):439-45. doi: 10.1093/gbe/evw002. Genome Biol Evol. 2016. PMID: 26798115 Free PMC article.
References
-
- Aliverti A, Pandini V, Pennati A, de Rosa M, Zanetti G. Structural and functional diversity of ferredoxin-NADP(+) reductases. Arch Biochem Biophys. 2008;474:283–291. - PubMed
-
- Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. - PubMed
-
- Marechal E, Cesbron-Delauw MF. The apicoplast: a new member of the plastid family. Trends Plant Sci. 2001;6:200–205. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

