BioRuby: bioinformatics software for the Ruby programming language
- PMID: 20739307
- PMCID: PMC2951089
- DOI: 10.1093/bioinformatics/btq475
BioRuby: bioinformatics software for the Ruby programming language
Abstract
Summary: The BioRuby software toolkit contains a comprehensive set of free development tools and libraries for bioinformatics and molecular biology, written in the Ruby programming language. BioRuby has components for sequence analysis, pathway analysis, protein modelling and phylogenetic analysis; it supports many widely used data formats and provides easy access to databases, external programs and public web services, including BLAST, KEGG, GenBank, MEDLINE and GO. BioRuby comes with a tutorial, documentation and an interactive environment, which can be used in the shell, and in the web browser.
Availability: BioRuby is free and open source software, made available under the Ruby license. BioRuby runs on all platforms that support Ruby, including Linux, Mac OS X and Windows. And, with JRuby, BioRuby runs on the Java Virtual Machine. The source code is available from http://www.bioruby.org/.
Contact: katayama@bioruby.org
Figures

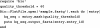
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

