Improved deoxyribozymes for synthesis of covalently branched DNA and RNA
- PMID: 20739352
- PMCID: PMC3017605
- DOI: 10.1093/nar/gkq753
Improved deoxyribozymes for synthesis of covalently branched DNA and RNA
Abstract
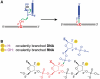
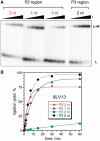
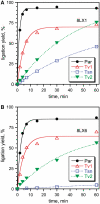
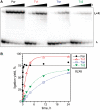
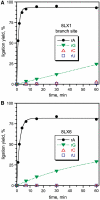
A covalently branched nucleic acid can be synthesized by joining the 2'-hydroxyl of the branch-site ribonucleotide of a DNA or RNA strand to the activated 5'-phosphorus of a separate DNA or RNA strand. We have previously used deoxyribozymes to synthesize several types of branched nucleic acids for experiments in biotechnology and biochemistry. Here, we report in vitro selection experiments to identify improved deoxyribozymes for synthesis of branched DNA and RNA. Each of the new deoxyribozymes requires Mn²(+) as a cofactor, rather than Mg²(+) as used by our previous branch-forming deoxyribozymes, and each has an initially random region of 40 rather than 22 or fewer combined nucleotides. The deoxyribozymes all function by forming a three-helix-junction (3HJ) complex with their two oligonucleotide substrates. For synthesis of branched DNA, the best new deoxyribozyme, 8LV13, has k(obs) on the order of 0.1 min⁻¹, which is about two orders of magnitude faster than our previously identified 15HA9 deoxyribozyme. 8LV13 also functions at closer-to-neutral pH than does 15HA9 (pH 7.5 versus 9.0) and has useful tolerance for many DNA substrate sequences. For synthesis of branched RNA, two new deoxyribozymes, 8LX1 and 8LX6, were identified with broad sequence tolerances and substantial activity at pH 7.5, versus pH 9.0 for many of our previous deoxyribozymes that form branched RNA. These experiments provide new, and in key aspects improved, practical catalysts for preparation of synthetic branched DNA and RNA.
Figures





References
-
- Chiuman W, Li Y. Making AppDNA using T4 DNA ligase. Bioorg. Chem. 2002;30:332–349. - PubMed
-
- Seeman NC. Biochemistry and structural DNA nanotechnology: an evolving symbiotic relationship. Biochemistry. 2003;42:7259–7269. - PubMed
-
- Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. - PubMed
-
- Lilley DM. Analysis of branched nucleic acid structure using comparative gel electrophoresis. Q. Rev. Biophys. 2008;41:1–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

