RNA reactions one molecule at a time
- PMID: 20739416
- PMCID: PMC2964187
- DOI: 10.1101/cshperspect.a003624
RNA reactions one molecule at a time
Abstract
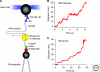
Much of the dynamics information is lost in bulk measurements because of the population averaging. Single-molecule methods measure one molecule at a time; they provide knowledge not obtainable by other means. In this article, we review the application of the two most widely used single-molecule methods--fluorescence resonance energy transfer (FRET) and force versus extension measurements--to several RNA reactions. First, we discuss folding/unfolding studies on a hairpin ribozyme that revealed multiple conformations of the RNA with distinct kinetics, and on a series of RNA pseudoknots, whose mechanical stabilities were found to show a strong correlation with their frameshifting efficiency during translation. We also discuss several RNA-related molecular motors. Single-molecule experiments revealed detailed mechanisms for the interaction of HIV reverse transcriptase and nucleic acid helicases (NS3 and RIG-1) with their substrates. Optical tweezers studies showed that translation of a single messenger RNA by a ribosome occurs by successive translocation-and-pause cycles. Single-molecule FRET experiments yielded important information on ribosome conformational changes and tRNA dynamics during translation. Overall, single-molecule experiments have been very valuable for understanding RNA reactions.
Figures


Similar articles
-
Single-Molecule Mechanical Folding and Unfolding of RNA Hairpins: Effects of Single A-U to A·C Pair Substitutions and Single Proton Binding and Implications for mRNA Structure-Induced -1 Ribosomal Frameshifting.J Am Chem Soc. 2018 Jul 5;140(26):8172-8184. doi: 10.1021/jacs.8b02970. Epub 2018 Jun 21. J Am Chem Soc. 2018. PMID: 29884019
-
How RNA unfolds and refolds.Annu Rev Biochem. 2008;77:77-100. doi: 10.1146/annurev.biochem.77.061206.174353. Annu Rev Biochem. 2008. PMID: 18518818 Review.
-
Translation at the single-molecule level.Annu Rev Biochem. 2008;77:177-203. doi: 10.1146/annurev.biochem.77.070606.101431. Annu Rev Biochem. 2008. PMID: 18518820 Review.
-
Ribosome structure and dynamics by smFRET microscopy.Methods Enzymol. 2014;549:375-406. doi: 10.1016/B978-0-12-801122-5.00016-7. Methods Enzymol. 2014. PMID: 25432757
-
Translocation kinetics and structural dynamics of ribosomes are modulated by the conformational plasticity of downstream pseudoknots.Nucleic Acids Res. 2018 Oct 12;46(18):9736-9748. doi: 10.1093/nar/gky636. Nucleic Acids Res. 2018. PMID: 30011005 Free PMC article.
Cited by
-
Mapping L1 ligase ribozyme conformational switch.J Mol Biol. 2012 Oct 12;423(1):106-22. doi: 10.1016/j.jmb.2012.06.035. Epub 2012 Jul 3. J Mol Biol. 2012. PMID: 22771572 Free PMC article.
-
The RNA worlds in context.Cold Spring Harb Perspect Biol. 2012 Jul 1;4(7):a006742. doi: 10.1101/cshperspect.a006742. Cold Spring Harb Perspect Biol. 2012. PMID: 21441585 Free PMC article.
-
Probing nucleic acid interactions and pre-mRNA splicing by Förster Resonance Energy Transfer (FRET) microscopy.Int J Mol Sci. 2012 Nov 14;13(11):14929-45. doi: 10.3390/ijms131114929. Int J Mol Sci. 2012. PMID: 23203103 Free PMC article. Review.
-
A guide to large-scale RNA sample preparation.Anal Bioanal Chem. 2018 May;410(14):3239-3252. doi: 10.1007/s00216-018-0943-8. Epub 2018 Mar 15. Anal Bioanal Chem. 2018. PMID: 29546546 Free PMC article. Review.
-
HIV-1 frameshift efficiency is primarily determined by the stability of base pairs positioned at the mRNA entrance channel of the ribosome.Nucleic Acids Res. 2013 Feb 1;41(3):1901-13. doi: 10.1093/nar/gks1254. Epub 2012 Dec 16. Nucleic Acids Res. 2013. PMID: 23248007 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
