Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing
- PMID: 20739521
- PMCID: PMC2953167
- DOI: 10.1128/JVI.01217-10
Tracking the evolution of multiple in vitro hepatitis C virus replicon variants under protease inhibitor selection pressure by 454 deep sequencing
Abstract
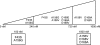
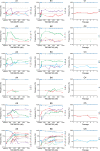
Resistance to hepatitis C virus (HCV) inhibitors targeting viral enzymes has been observed in in vitro replicon studies and during clinical trials. The factors determining the emergence of resistance and the changes in the viral quasispecies population under selective pressure are not fully understood. To assess the dynamics of variants emerging in vitro under various selective pressures with TMC380765, a potent macrocyclic HCV NS3/4A protease inhibitor, HCV genotype 1b replicon-containing cells were cultured in the presence of a low, high, or stepwise-increasing TMC380765 concentration(s). HCV replicon RNA from representative samples thus obtained was analyzed using (i) population, (ii) clonal, and (iii) 454 deep sequencing technologies. Depending on the concentration of TMC380765, distinct mutational patterns emerged. In particular, culturing with low concentrations resulted in the selection of low-level resistance mutations (F43S and A156G), whereas high concentrations resulted in the selection of high-level resistance mutations (A156V, D168V, and D168A). Clonal and 454 deep sequencing analysis of the replicon RNA allowed the identification of low-frequency preexisting mutations possibly contributing to the mutational pattern that emerged. Stepwise-increasing TMC380765 concentrations resulted in the emergence and disappearance of multiple replicon variants in response to the changing selection pressure. Moreover, two different codons for the wild-type amino acids were observed at certain NS3 positions within one population of replicons, which may contribute to the emerging mutational patterns. Deep sequencing technologies enabled the study of minority variants present in the HCV quasispecies population present at baseline and during antiviral drug pressure, giving new insights into the dynamics of resistance acquisition by HCV.
Figures




References
-
- Adiwijaya, B. S., E. Herrmann, B. Hare, T. Kieffer, C. Lin, A. D. Kwong, V. Garg, J. C. Randle, C. Sarrazin, S. Zeuzem, and P. R. Caron. 2010. A multi-variant, viral dynamic model of genotype 1 HCV to assess the in vivo evolution of protease-inhibitor resistant variants. PLoS Comput. Biol. 6:e1000745. - PMC - PubMed
-
- Bartels, D. J., Y. Zhou, E. Z. Zhang, M. Marcial, R. A. Byrn, T. Pfeiffer, A. M. Tigges, B. S. Adiwijaya, C. Lin, A. D. Kwong, and T. L. Kieffer. 2008. Natural prevalence of hepatitis C virus variants with decreased sensitivity to NS3.4A protease inhibitors in treatment-naive subjects. J. Infect. Dis. 198:800-807. - PubMed
-
- Biebricher, C. K., and M. Eigen. 2005. The error threshold. Virus Res. 107:117-127. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

