Novel acid phosphatase in Candida glabrata suggests selective pressure and niche specialization in the phosphate signal transduction pathway
- PMID: 20739710
- PMCID: PMC2975289
- DOI: 10.1534/genetics.110.120824
Novel acid phosphatase in Candida glabrata suggests selective pressure and niche specialization in the phosphate signal transduction pathway
Abstract
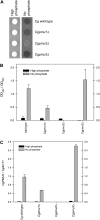
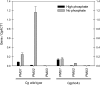
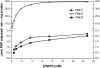
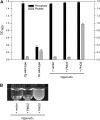
Evolution through natural selection suggests unnecessary genes are lost. We observed that the yeast Candida glabrata lost the gene encoding a phosphate-repressible acid phosphatase (PHO5) present in many yeasts including Saccharomyces cerevisiae. However, C. glabrata still had phosphate starvation-inducible phosphatase activity. Screening a C. glabrata genomic library, we identified CgPMU2, a member of a three-gene family that contains a phosphomutase-like domain. This small-scale gene duplication event could allow for sub- or neofunctionalization. On the basis of phylogenetic and biochemical characterizations, CgPMU2 has neofunctionalized to become a broad range, phosphate starvation-regulated acid phosphatase, which functionally replaces PHO5 in this pathogenic yeast. We determined that CgPmu2, unlike ScPho5, is not able to hydrolyze phytic acid (inositol hexakisphosphate). Phytic acid is present in fruits and seeds where S. cerevisiae grows, but is not abundant in mammalian tissues where C. glabrata grows. We demonstrated that C. glabrata is limited from an environment where phytic acid is the only source of phosphate. Our work suggests that during evolutionary time, the selection for the ancestral PHO5 was lost and that C. glabrata neofunctionalized a weak phosphatase to replace PHO5. Convergent evolution of a phosphate starvation-inducible acid phosphatase in C. glabrata relative to most yeast species provides an example of how small changes in signal transduction pathways can mediate genetic isolation and uncovers a potential speciation gene.
Figures



Similar articles
-
A paralogue of the phosphomutase-like gene family in Candida glabrata, CgPmu2, gained broad-range phosphatase activity due to a small number of clustered substitutions.Biochem J. 2015 Oct 15;471(2):187-98. doi: 10.1042/BJ20150611. Epub 2015 Aug 12. Biochem J. 2015. PMID: 26268557
-
Dynamic Changes in Yeast Phosphatase Families Allow for Specialization in Phosphate and Thiamine Starvation.G3 (Bethesda). 2018 Jul 2;8(7):2333-2343. doi: 10.1534/g3.118.200303. G3 (Bethesda). 2018. PMID: 29748198 Free PMC article.
-
Candida glabrata PHO4 is necessary and sufficient for Pho2-independent transcription of phosphate starvation genes.Genetics. 2009 Jun;182(2):471-9. doi: 10.1534/genetics.109.101063. Epub 2009 Mar 30. Genetics. 2009. PMID: 19332882 Free PMC article.
-
Multiple interfaces control activity of the Candida glabrata Pdr1 transcription factor mediating azole drug resistance.Curr Genet. 2019 Feb;65(1):103-108. doi: 10.1007/s00294-018-0870-4. Epub 2018 Jul 28. Curr Genet. 2019. PMID: 30056490 Free PMC article. Review.
-
The mating type-like loci of Candida glabrata.Rev Iberoam Micol. 2014 Jan-Mar;31(1):30-4. doi: 10.1016/j.riam.2013.09.016. Epub 2013 Nov 16. Rev Iberoam Micol. 2014. PMID: 24252826 Review.
Cited by
-
Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice.Antimicrob Agents Chemother. 2011 May;55(5):1852-60. doi: 10.1128/AAC.01271-10. Epub 2011 Feb 14. Antimicrob Agents Chemother. 2011. PMID: 21321146 Free PMC article.
-
Phosphate in Virulence of Candida albicans and Candida glabrata.J Fungi (Basel). 2020 Mar 26;6(2):40. doi: 10.3390/jof6020040. J Fungi (Basel). 2020. PMID: 32224872 Free PMC article. Review.
-
A Novel cis Element Achieves the Same Solution as an Ancestral cis Element During Thiamine Starvation in Candida glabrata.G3 (Bethesda). 2020 Jan 7;10(1):321-331. doi: 10.1534/g3.119.400897. G3 (Bethesda). 2020. PMID: 31732505 Free PMC article.
-
Core regulatory components of the PHO pathway are conserved in the methylotrophic yeast Hansenula polymorpha.Curr Genet. 2016 Aug;62(3):595-605. doi: 10.1007/s00294-016-0565-7. Epub 2016 Jan 21. Curr Genet. 2016. PMID: 26794724 Free PMC article.
-
Comparative genomics of emerging pathogens in the Candida glabrata clade.BMC Genomics. 2013 Sep 14;14:623. doi: 10.1186/1471-2164-14-623. BMC Genomics. 2013. PMID: 24034898 Free PMC article.
References
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne and S. Brunak, 2004. Improved prediction of signal peptides: signalP 3.0. J. Mol. Biol. 340 783–795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

