Structured digital tables on the Semantic Web: toward a structured digital literature
- PMID: 20739925
- PMCID: PMC2950080
- DOI: 10.1038/msb.2010.45
Structured digital tables on the Semantic Web: toward a structured digital literature
Abstract
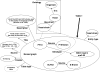
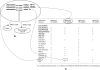
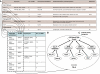
In parallel to the growth in bioscience databases, biomedical publications have increased exponentially in the past decade. However, the extraction of high-quality information from the corpus of scientific literature has been hampered by the lack of machine-interpretable content, despite text-mining advances. To address this, we propose creating a structured digital table as part of an overall effort in developing machine-readable, structured digital literature. In particular, we envision transforming publication tables into standardized triples using Semantic Web approaches. We identify three canonical types of tables (conveying information about properties, networks, and concept hierarchies) and show how more complex tables can be built from these basic types. We envision that authors would create tables initially using the structured triples for canonical types and then have them visually rendered for publication, and we present examples for converting representative tables into triples. Finally, we discuss how 'stub' versions of structured digital tables could be a useful bridge for connecting together the literature with databases, allowing the former to more precisely document the later.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Berners-Lee T, Hendler J, Lassila O (2001) The Semantic Web. Sci Am 284: 34–43 - PubMed
-
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball C, Causton H, Gaasterland T, Glenisson P, Holstege F, Kim I, Markowitz V, Matese J, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S et al. (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet 29: 365–371 - PubMed
-
- Carroll J, Bizer C, Hayes P, Stickler P (2005) Named graphs. Web Semant 3: 247–267
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

