The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres
- PMID: 20739937
- PMCID: PMC2946842
- DOI: 10.1038/nature09323
The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres
Abstract
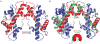
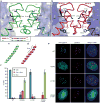
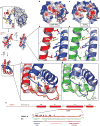
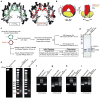
Centromeres are specified epigenetically, and the histone H3 variant CENP-A is assembled into the chromatin of all active centromeres. Divergence from H3 raises the possibility that CENP-A generates unique chromatin features to mark physically centromere location. Here we report the crystal structure of a subnucleosomal heterotetramer, human (CENP-A-H4)(2), that reveals three distinguishing properties encoded by the residues that comprise the CENP-A targeting domain (CATD; ref. 2): (1) a CENP-A-CENP-A interface that is substantially rotated relative to the H3-H3 interface; (2) a protruding loop L1 of the opposite charge as that on H3; and (3) strong hydrophobic contacts that rigidify the CENP-A-H4 interface. Residues involved in the CENP-A-CENP-A rotation are required for efficient incorporation into centromeric chromatin, indicating specificity for an unconventional nucleosome shape. DNA topological analysis indicates that CENP-A-containing nucleosomes are octameric with conventional left-handed DNA wrapping, in contrast to other recent proposals. Our results indicate that CENP-A marks centromere location by restructuring the nucleosome from within its folded histone core.
Figures
References
-
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. - PubMed
-
- Black BE, et al. Structural determinants for generating centromeric chromatin. Nature. 2004;430:578–582. - PubMed
-
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

