The Role of Lipids in Retrovirus Replication
- PMID: 20740061
- PMCID: PMC2927015
- DOI: 10.3390/v2051146
The Role of Lipids in Retrovirus Replication
Abstract
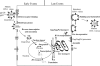
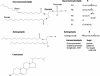
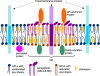
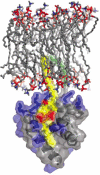
Retroviruses undergo several critical steps to complete a replication cycle. These include the complex processes of virus entry, assembly, and budding that often take place at the plasma membrane of the host cell. Both virus entry and release involve membrane fusion/fission reactions between the viral envelopes and host cell membranes. Accumulating evidence indicates important roles for lipids and lipid microdomains in virus entry and egress. In this review, we outline the current understanding of the role of lipids and membrane microdomains in retroviral replication.
Figures


Similar articles
-
Lipids and membrane microdomains in HIV-1 replication.Virus Res. 2009 Aug;143(2):162-76. doi: 10.1016/j.virusres.2009.04.007. Epub 2009 Apr 19. Virus Res. 2009. PMID: 19383519 Free PMC article. Review.
-
Virus infection and lipid rafts.Biol Pharm Bull. 2006 Aug;29(8):1538-41. doi: 10.1248/bpb.29.1538. Biol Pharm Bull. 2006. PMID: 16880600 Review.
-
Role of lipids in virus replication.Cold Spring Harb Perspect Biol. 2011 Oct 1;3(10):a004820. doi: 10.1101/cshperspect.a004820. Cold Spring Harb Perspect Biol. 2011. PMID: 21628428 Free PMC article. Review.
-
Host Cell Plasma Membrane Phosphatidylserine Regulates the Assembly and Budding of Ebola Virus.J Virol. 2015 Sep;89(18):9440-53. doi: 10.1128/JVI.01087-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136573 Free PMC article.
-
Assembly and budding of influenza virus.Virus Res. 2004 Dec;106(2):147-65. doi: 10.1016/j.virusres.2004.08.012. Virus Res. 2004. PMID: 15567494 Free PMC article. Review.
Cited by
-
How HIV finds the door.Proc Natl Acad Sci U S A. 2012 Nov 13;109(46):18631-2. doi: 10.1073/pnas.1215940109. Epub 2012 Nov 1. Proc Natl Acad Sci U S A. 2012. PMID: 23118338 Free PMC article. No abstract available.
-
The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization.J Biol Chem. 2020 Jan 10;295(2):348-362. doi: 10.1074/jbc.RA119.009481. Epub 2019 Nov 22. J Biol Chem. 2020. PMID: 31757809 Free PMC article.
-
Global rescue of defects in HIV-1 envelope glycoprotein incorporation: implications for matrix structure.PLoS Pathog. 2013;9(11):e1003739. doi: 10.1371/journal.ppat.1003739. Epub 2013 Nov 14. PLoS Pathog. 2013. PMID: 24244165 Free PMC article.
-
New insights into HIV assembly and trafficking.Physiology (Bethesda). 2011 Aug;26(4):236-51. doi: 10.1152/physiol.00051.2010. Physiology (Bethesda). 2011. PMID: 21841072 Free PMC article. Review.
-
Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers.Proc Natl Acad Sci U S A. 2013 Feb 26;110(9):3525-30. doi: 10.1073/pnas.1216655110. Epub 2013 Feb 11. Proc Natl Acad Sci U S A. 2013. PMID: 23401539 Free PMC article.
References
-
- Ono A, Freed EO. Role of lipid rafts in virus replication. Adv Virus Res. 2005;64:311–358. - PubMed
-
- Freed E, Martin M. HIVs and their replication. In: Knipe D, Howley P, editors. Fields Virology. Lippincott; Williams, and Wilkins, USA: 2007. pp. 2107–2185.
-
- Munro S. Lipid rafts: elusive or illusive. Cell. 2003;115:377–388. - PubMed
-
- Simons K, Vaz WL. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct. 2004;33:269–295. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
