Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents
- PMID: 20740281
- PMCID: PMC3011088
- DOI: 10.1007/s00248-010-9735-1
Recombination within and between species of the alpha proteobacterium Bartonella infecting rodents
Abstract
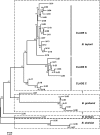
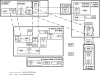
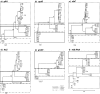
Bartonella infections from wild mice and voles (Apodemus flavicollis, Mi. oeconomus, Microtus arvalis and Myodes glareolus) were sampled from a forest and old-field habitats of eastern Poland; a complex network of Bartonella isolates, referrable to B. taylorii, B. grahamii, B. birtlesii and B. doshiae, was identified by the sequencing of a gltA fragment, comparable to previous studies of Bartonella diversity in rodents. Nested clade analysis showed that isolates could be assigned to zero- and one-step clades which correlated with host identity and were probably the result of clonal expansion; however, sequencing of other housekeeping genes (rpoB, ribC, ftsZ, groEl) and the 16S RNA gene revealed a more complex situation with clear evidence of numerous recombinant events in which one or both Bartonella parents could be identified. Recombination within gltA was found to have generated two distinct variant clades, one a hybrid between B. taylorii and B. doshiae, the other between B. taylorii and B. grahamii. These recombinant events characterised the differences between the two-step and higher clades within the total nested cladogram, involved all four species of Bartonella identified in this work and appear to have played a dominant role in the evolution of Bartonella diversity. It is clear, therefore, that housekeeping gene phylogenies are not robust indicators of Bartonella diversity, especially when only a single gene (gltA or 16S RNA) is used. Bartonella clades infecting Microtus were most frequently involved in recombination and were most frequently tip clades within the cladogram. The role of Microtus in influencing the frequency of Bartonella recombination remains unknown.
Figures

References
-
- Berglund EC, Frank AC, Calteau A, Petterson OV, Granberg F, Eriksson AS, Naslund K, Holmberg M, Lindroos H, Andersson SGE. Run off replication of host adaptability genes is associated with gene transfer agents in the genome of mouse infecting Bartonella grahamii. PLoS Genet. 2009;5:e1000546. doi: 10.1371/journal.pgen.1000546. - DOI - PMC - PubMed
-
- Birtles RJ, Harrison TG, Molyneux DH. Grahamella in small woodland mammals in the UK—isolation, prevalence and host specificity. Ann Trop Med Parasitol. 1994;88:317–327. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

