Collagen oligomers modulate physical and biological properties of three-dimensional self-assembled matrices
- PMID: 20740490
- PMCID: PMC3387932
- DOI: 10.1002/bip.21537
Collagen oligomers modulate physical and biological properties of three-dimensional self-assembled matrices
Abstract
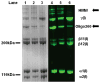
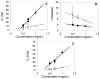
Elucidation of mechanisms underlying collagen fibril assembly and matrix-induced guidance of cell fate will contribute to the design and expanded use of this biopolymer for research and clinical applications. Here, we define how Type I collagen oligomers affect in-vitro polymerization kinetics as well as fibril microstructure and mechanical properties of formed matrices. Monomers and oligomers were fractionated from acid-solubilized pig skin collagen and used to generate formulations varying in monomer/oligomer content or average polymer molecular weight (AMW). Polymerization half-times decreased with increasing collagen AMW and closely paralleled lag times, indicating that oligomers effectively served as nucleation sites. Furthermore, increasing AMW yielded matrices with increased interfibril branching and had no correlative effect on fibril density or diameter. These microstructure changes increased the stiffness of matrices as evidenced by increases in both shear storage and compressive moduli. Finally, the biological relevance of modulating collagen AMW was evidenced by the ability of cultured endothelial colony forming cells to sense associated changes in matrix physical properties and alter vacuole and capillary-like network formation. This work documents the importance of oligomers as another physiologically-relevant design parameter for development and standardization of polymerizable collagen formulations to be used for cell culture, regenerative medicine, and engineered tissue applications.
© 2010 Wiley Periodicals, Inc.
Figures








References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

