Adhesion through single peptide aptamers
- PMID: 20795685
- PMCID: PMC4047793
- DOI: 10.1021/jp1031493
Adhesion through single peptide aptamers
Abstract
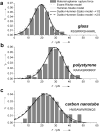
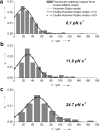
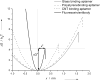
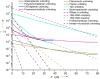
Aptamer and antibody mediated adhesion is central to biological function and is valuable in the engineering of "lab on a chip" devices. Single molecule force spectroscopy using optical tweezers enables direct nonequilibrium measurement of these noncovalent interactions for three peptide aptamers selected for glass, polystyrene, and carbon nanotubes. A comprehensive examination of the strong attachment between antifluorescein 4-4-20 and fluorescein was also carried out using the same assay. Bond lifetime, barrier width, and free energy of activation are extracted from unbinding histogram data using three single molecule pulling models. The evaluated aptamers appear to adhere stronger than the fluorescein antibody under no- and low-load conditions, yet weaker than antibodies at loads above ∼25 pN. Comparison to force spectroscopy data of other biological linkages shows the diversity of load dependent binding and provides insight into linkages used in biological processes and those designed for engineered systems.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

